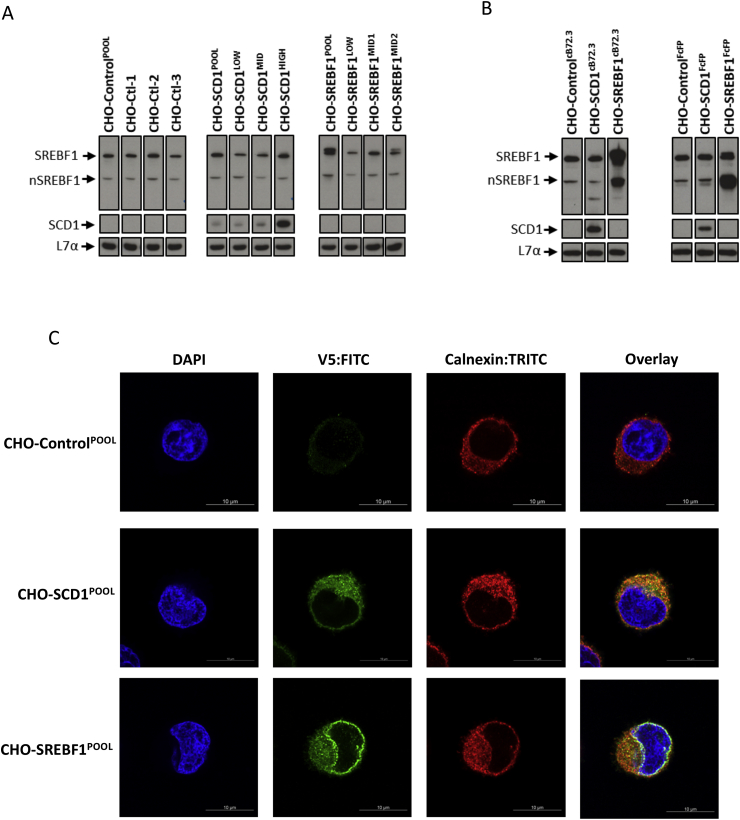
Fig. 2.
Overexpression and cellular localization of SCD1 and SREBF1 in engineered cells. Western blot analysis of CHOK1SV GS-KO™ cells engineered to overexpress either SCD1 or SREBF1 genes where controls have been generated with an unmodified pcDNA3.1V5-His/TOPO vector. CHO-Ctl-1, CHO-Ctl-2 and CHO-Ctl-3 are monoclonal lines derived from the CHO-ControlPOOL; CHO-SCD1LOW, CHO-SCD1MID and CHO-SCD1HIGH are monoclonal cell lines isolated from CHO-SCD1POOL and CHO-SREBF1LOW, CHO-SREBF1MID1 and CHO-SREBF1MID2 are monoclonal lines isolated from CHO-SREBF1POOL. The pools which had been engineered were used to generate cells expressing cB72.3 (CHO-ControlcB72.3, CHO-SCD1cB72.3 and CHO-SREBF1cB72.3) or an Fc fusion protein (FcFP; CHO-ControlFcFP, CHO-SCD1FcFP and CHO-SREBF1FcFP). Lysate samples were generated from these cells harvested at day 6 of culture from either host cells (A) or cells stably producing the recombinant products cB72.3 or FcFP where specified (B). A primary antibody with specificity for L7α was used as a loading control. (C) Cellular localization of overexpressed SCD1 and SREBF1 proteins in CHO-ControlPOOL, CHO-SCD1POOL and CHO-SREBF1POOL cell pools as determined by immunofluorescent detection using an anti-V5 antibody conjugated with a FITC secondary antibody. An anti-calnexin antibody conjugated with a TRITC secondary antibody was used to highlight the position of the ER. Images were obtained using confocal microscopy (see Data in Brief (Budge et al., 2019) Table 2 for full details of antibodies used).