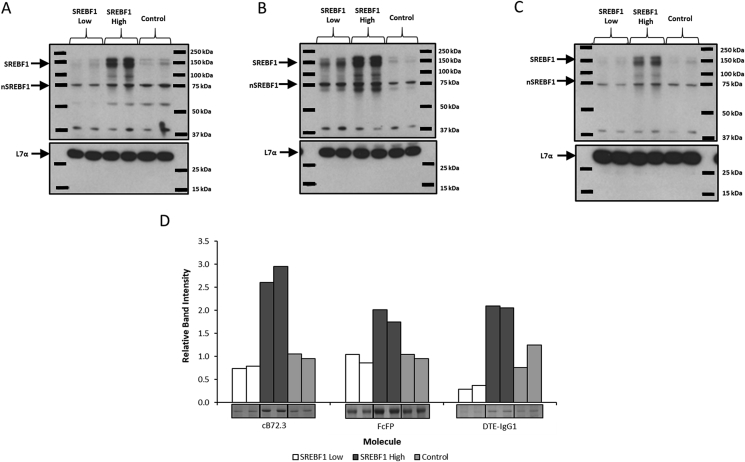
Fig. 5.
Analysis of SREBF1 engineered CHOK1SV GS-KO™ host cells stably expressing a variety of secretory recombinant biotherapeutic molecules. Cell pools were constructed by the simultaneous co-transfection of two expression vectors. The first vector contained the GS gene for selection as well as genes appropriate for production of either cB72.3, FcFP or DTE-IgG1 whilst the second vector contained the SREBF1 gene driven by different promoters to yield either low (SREBF1 Low) or high (SREBF1 High) SREBF1 protein abundance. Control pools (Control) were generated using the appropriate first vector (also generated using GS selection) in conjunction with an empty second vector (i.e. not encoding the SREBF1 gene). Western blot analysis was carried out to determine relative levels of SREBF1 abundance from cell pools constructed to express either cB72.3 (A), FcFP (B) or DTE-IgG1 (C) where SREBF1 Low, SREBF1 High or a Control vector were co-transfected during the generation of pools as highlighted in the figure (L7a was used as a loading control). Figure D shows bands from a Coomassie blue stained SDS-PAGE gel and densitometry analysis of bands corresponding to the recombinant products harvested at 7 days of batch culture from the constructed cell pools. All lanes show bands corresponding to an independent cell pool construction.