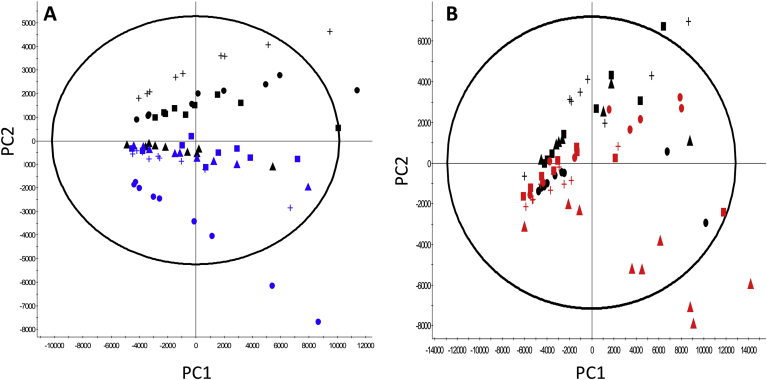
Fig. 6.
Analysis of cellular lipid content in control and LMM engineered CHO cells using mass spectrometry. (A) and (B) show Principal Component Analysis (PCA) of the mass spectrometry derived lipid profiles extracted from cells overexpressing either SREBF1 or SCD1 genes highlighting differences in lipidomic profiles of engineered CHOK1SV GS-KO cell lines harvested after 6 days of batch culture. Figure A shows PCA of control samples and samples engineered to overexpress SCD1; black data points represent controls (CHO-Ctl-1 = ●, CHO-Ctl-2 = ▲, CHO-Ctl-3 = +, CHO-ControlPOOL = ■) whilst blue data points represent SCD1 overexpressing cells (CHO-SCD1LOW =  , CHO-SCD1MID = +, CHO-SCD1HIGH =
, CHO-SCD1MID = +, CHO-SCD1HIGH =  , CHO-SCD1POOL =
, CHO-SCD1POOL =  ). Figure B shows analysis of control samples and samples engineered to overexpress SREBF1; black data points represent controls (CHO-Ctl-1 = ●, CHO-Ctl-2 = ▲, CHO-Ctl-3 = +, CHO-ControlPOOL = ■) whilst red data points represent SREBF1 overexpressing cells (CHO-SREBF1LOW =
). Figure B shows analysis of control samples and samples engineered to overexpress SREBF1; black data points represent controls (CHO-Ctl-1 = ●, CHO-Ctl-2 = ▲, CHO-Ctl-3 = +, CHO-ControlPOOL = ■) whilst red data points represent SREBF1 overexpressing cells (CHO-SREBF1LOW =  , CHO-SREBF1MID1 =
, CHO-SREBF1MID1 =  , CHO-SREBF1MID2 =
, CHO-SREBF1MID2 =  , CHO-SREBF1POOL =
, CHO-SREBF1POOL =  ). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)