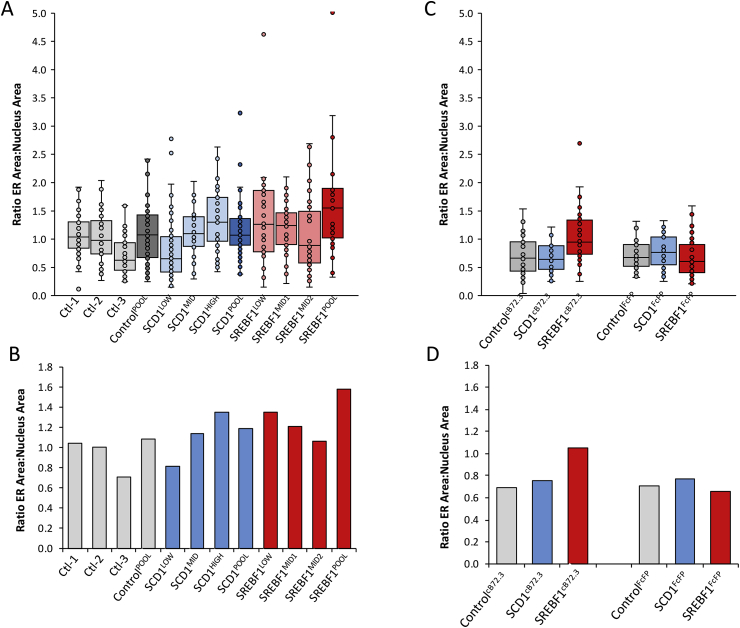
Fig. 8.
Quantification of ER area in relation to nuclear area calculated using immunofluorescence confocal microscopy. ER and nuclear features which were highlighted using an anti-calnexin antibody (an established ER marker (Roobol et al., 2009)) followed by conjugation with a TRITC secondary antibody and DAPI respectively and images were taken at the approximate equator of the cell on the z axis, as determined by the intensity and diameter of the DAPI stain. Values obtained were measured using the “spline contour” tool of Zen blue software to outline the ER and nuclear features of the cells. All cells were adhered and fixed to coverslips on day 6 of batch culture and all images analyzed were from this timepoint. A & B show values from pools and monoclonal host CHO cells engineered to overexpress SCD1 or SREBF1 and control CHO cells, generated using the original pcDNA3.1V5-His/TOPO vector whilst C & D show cells stably expressing either cB72.3 or the FcFP generated from control, SCD1 or SREBF1 host pools. Further, A & C show box and whisker plots generated from values obtained where all individual recorded data points are represented by dots and B & D show mean values obtained using these data points (n ≥ 30). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)