Abstract
Differently from other myeloid cells, microglia derive exclusively from precursors originating within the yolk sac and migrate to the CNS under development, without any contribution from fetal liver or postnatal hematopoiesis. Consistent with their unique ontology, microglia may express specific physiological markers, which have been partly described in recent years. Here we wondered whether profiles distinguishing microglia from peripheral macrophages vary with age and under pathology. To this goal, we profiled transcriptomes of microglia throughout the lifespan and included a parallel comparison with peripheral macrophages under physiological and neuroinflammatory settings using age- and sex-matched wild-type and bone marrow chimera mouse models. This comprehensive approach demonstrated that the phenotypic differentiation between microglia and peripheral macrophages is age-dependent and that peripheral macrophages do express some of the most commonly described microglia-specific markers early during development, such as Fcrls, P2ry12, Tmem119, and Trem2. Further, during chronic neuroinflammation CNS-infiltrating macrophages and not peripheral myeloid cells acquire microglial markers, indicating that the CNS niche may instruct peripheral myeloid cells to gain the phenotype and, presumably, the function of the microglia cell. In conclusion, our data provide further evidence about the plasticity of the myeloid cell and suggest caution in the strict definition and application of microglia-specific markers.
SIGNIFICANCE STATEMENT Understanding the respective role of microglia and infiltrating monocytes in neuroinflammatory conditions has recently seemed possible by the identification of a specific microglia signature. Here instead we provide evidence that peripheral macrophages may express some of the most commonly described microglia markers at some developmental stages or pathological conditions, in particular during chronic neuroinflammation. Further, our data support the hypothesis about phenotypic plasticity and convergence among distinct myeloid cells so that they may act as a functional unit rather than as different entities, boosting their mutual functions in different phases of disease. This holds relevant implications in the view of the growing use of myeloid cell therapies to treat brain disease in humans.
Keywords: development, EAE, macrophages, microglia, plasticity, tissue-reprogramming
Introduction
Microglia are the resident macrophages of the brain and play vital functions for CNS development and homeostasis (Salter and Stevens, 2017; Li and Barres, 2018). They are long-lived cells deriving from a transient hematopoietic wave of erythro-myeloid progenitors emerging in the yolk sac (YS; Li and Barres, 2018). Unlike other tissues, however, the CNS does not receive any further precursors from fetal liver or postnatal hematopoiesis due to the maturation of the blood–brain barrier (at E14 in the mouse), so that microglia remain the exclusively YS-derived myeloid cells in the CNS throughout life (Ginhoux et al., 2010; Schulz et al., 2012; Kierdorf et al., 2013; Gomez Perdiguero et al., 2015; Hoeffel and Ginhoux, 2015; Hagemeyer et al., 2017). In the view of their unique ontology, many efforts have been made to identify microglia-specific markers to be used for their selective manipulation (Gautier et al., 2012; Hickman et al., 2013; Butovsky et al., 2014; Zhang et al., 2014). Recent literature has shed some light into the complexity of microglial phenotype, showing that it is constantly shaped by the microenvironment in a time- and region-dependent manner (Gosselin et al., 2014, 2017; Lavin et al., 2014; Mass et al., 2016; Matcovitch-Natan et al., 2016; Hammond et al., 2019; Masuda et al., 2019), however whether this phenotype remains distinct from peripheral myeloid cells throughout the lifespan and under pathological conditions needs to be clarified. To this goal, we performed whole-genome transcriptome studies on brain microglia and peripheral macrophages in three critical time windows, i.e., at the mist of embryonic life (E14.5), in the early postnatal life (P1) and in adulthood (P60). These time points have been chosen as paradigmatic periods during which microglia display neurosupportive functions, such as control of the proper brain development during embryonic life (Antony et al., 2011; Cunningham et al., 2013; Ueno et al., 2013; Arnò et al., 2014; Squarzoni et al., 2014), maturation of a full-functional synaptic network soon after birth (Paolicelli et al., 2011; Derecki et al., 2012; Schafer and Stevens, 2015) and finally regulation of brain homeostasis in adulthood (Hanisch and Kettenmann, 2007; Prinz et al., 2011; Salter and Stevens, 2017). This approach led to the identification of novel time-dependent microglial signatures and of a core of stable microglia-specific transcripts throughout life. A selected subset of microglial markers was then monitored on microglia and peripheral myeloid cells during experimental neuroinflammation in wild-type mice and bone-marrow chimera. These experiments demonstrated that under pathological settings CNS-infiltrating and not peripheral macrophages may acquire even the most specific microglia markers, thus indicating the instructive action of the CNS niche in shaping phenotypic convergence between infiltrating and resident myeloid cells.
Materials and Methods
Animals.
C57BL/6 mice were purchased from Charles River Laboratories. All efforts were made to minimize animal suffering and reduce the number of mice used in accordance with the European Communities Council Directive of November 24, 1986 (86/609/EEC). All animal experimental protocols were approved by the Ethics Review Committee for Animal Experimentation of the Italian Ministry of Health. Procedures were performed according to the guidelines of the Institutional Animal Care and Use Committee of the San Raffaele Scientific Institute (protocol 727/2011).
Cell preparation and sorting.
For the preparation of embryonic and postnatal samples, cell suspensions were made from brain and liver of 18–20 C57BL/6 E14.5/15.5 embryos or P1 pups. Tissues were cut into small pieces and incubated in a solution containing ice-cold 1× HBSS, 0.6% glucose, 5 mm HEPES, pH 7.4. Cell suspensions were filtered through 40 μm cell strainer, then washed three times in 1× HBSS. Red blood cell lysis was performed on liver samples.
For the preparation of microglia from the CNS of adult naive mice, 2-month-old C57BL6 female mice were transcardially perfused with ice-cold PBS, and brains were extracted, 10 total brains were pooled, cut into small pieces, incubated for 30 min with 0.2 mg/ml type IV collagenase (Sigma-Aldrich), and afterward passed through a 40 μm cell strainer to obtain homogeneous cell suspensions. These preparations were centrifuged by a 37%/70% discontinuous Percoll gradient (GE Healthcare), and microglia cells were isolated from the interface (Cardona et al., 2006).
Isolated cells were stained with the fluorochrome-conjugated mAbs specific for mouse CD11b (clone M1/70) and CD45 (clone 30-F11) and with propidium iodide (PI; BD Biosciences) for vitality. Microglia cells were sorted based on the live (PI-negative) cell gate and the CD11b high/CD45 low expression profile; liver macrophages were sorted based on the live (PI-negative) cell gate and the CD11b high/CD45 high expression profile. Cells were sorted straight into the lysis buffer for RNA extraction. FACS sorting was performed on BD FACSAria III (Becton Dickinson).
EAE and bone marrow chimera.
Chronic experimental autoimmune encephalomyelitis (EAE) was induced in 6- to 8-week-old C57BL/6 female mice (Charles River Laboratories) by subcutaneous immunization with MOG35-55 peptide (200 mg per mouse) in incomplete Freund's adjuvant containing 4 mg/ml mycobacterium tuberculosis. Mice were intravenously injected with 500 ng of pertussis toxin (Quadratech) the day of the immunization and 2 d later [i.e., 0 and 2 d post-immunization (dpi)]. The animals were assessed daily for bodyweight and clinical symptoms of EAE. Their disease stage was recorded with the following scoring system: 0, healthy; 1, limp tail; 1.5, wobbling gait; 2, slight paresis of hind limbs/difficulty in righting; 2.5, severe paralysis of hind limbs; 3, complete paralysis of hind limbs; 3.5, initial paresis of forelimbs; 4, tetraparalysis; and 5, death.
For the generation of bone marrow chimera, 6- to 8-week-old C57BL/6 female mice were used as recipients, whereas donor mice were 4- to 6-week-old transgenic UBI-GFP/BL6 female mice (Schaefer et al., 2001). Recipient mice were sub-lethally exposed to a single dose of 1100 R from a Cesium source before transplantation. Mouse bone marrow cells were harvested from UBIGFP/BL6 mice by flushing femurs and tibiae. Then, cells (2 × 106 cells/mouse) were injected into the tail vein of recipient female. Two mounts later, 100 μl of total blood was collected from the tail vein of bone marrow transplanted mice and blood leukocytes were analyzed for GFP detection by flow cytometry. Only those mice with a percentage of chimerism >70% underwent EAE induction, as described at the beginning of this section.
EAE wild-type as well as GFP chimeric mice were killed at three different time points, i.e., pre-onset (8–9 dpi; median clinical score: 0), peak of disease (16–18 dpi; median clinical score: 2.8), and chronic phase (38–40 dpi; median clinical score: 2.2). At each time point they were killed by anesthetic overdose and transcardially perfused with ice-cold PBS. Brains were dissected and 10 of them were pooled. Microglia and infiltrating macrophages were isolated as described in the previous paragraph.
Genome-wide expression analysis.
Total RNA isolation was performed with RNeasy Micro kit (Qiagen). Pure RNA was eluted from the column in RNase-free water. RNA samples were kept at −80°C until use. Quality of the RNA samples was determined at NanoDrop spectrophotometer (ThermoFisher Scientific). RNA quality was confirmed with a 2100 Bioanalyzer (Agilent). Only samples having RNA Integrity Index >8 were used. Total RNA was subjected to amplification with an Illumina TotalPrep RNA Amplification Kit (Applied Biosystems). cRNA quality and quantity was again assessed at NanoDrop and by Experion RNA StdSens quality control kit (Bio-Rad). A total amount of 500 ng cRNA was hybridized to Illumina MouseRef-8 v2.0 Expression BeadChips, slides were processed according to the manufacturer's instructions and scanned with iScan (Illumina). Raw data were subjected to background subtraction followed by cubic spline normalization using Genome Studio software (Illumina). Filtering was performed for probes with detection p < 0.05 in at least 40% of samples. Principal component analysis and hierarchical sample cluster revealed three outlier samples in the E14 Brain group, which were removed from the subsequent analyses. For the detection of differentially expressed genes, empirical Bayes tests were performed as implemented in the LIMMA package in R-bioconductor platform. The significance threshold was set to Benjamini–Hochberg's corrected p < 0.01 for the intra-tissue comparison and p < 0.05 in case of inter-tissue comparison. The differentially expressed genes (DEGs) were defined by statistical significance, thresholds on fold-change (±1.7) and expression intensity (>100 in any 1 of the 2 compared groups). Gene ontology (GO) enrichment analysis and pathway analysis were performed using the Metacore suite (Thompson Reuters) or ToppFun in ToppGene suite (https://toppgene.cchmc.org), with the enrichment criteria of p < 0.05 after hypergeometric test followed by Benjamini–Hochberg's correction. The transcriptomics datasets generated in this study were uploaded in ArrayExpress database (E-MTAB-8059), whereas the transcriptomics dataset relative to unstimulated, M1 and M2 polarized mouse macrophages was described by Garzetti et al. (2014) and Colombo et al. (2018) and recovered from ArrayExpress database (accession no. E-MTAB-6416). Clustering analysis was performed by Spearman's rank correlation coefficient distance matrix with average linkage and heatmaps were generated using GeneE platform analysis tool (http://www.broadinstitute.org/cancer/software/GENE-E/). Principal component analysis (PCA) was performed in R using pca3D package.
Targeted molecular validation.
Total RNA extraction from cells isolated from EAE mice, including chimeric mice, were performed as described in the previous paragraph. Each experimental group consisted of two independent biological replicates derived from the pool of two samples. Each sample was derived from a pool of 10 brain and spinal cords. Total RNA (200 ng) was used in 40 μl of reverse transcription reaction (Superscript VILO cDNA Synthesis Kit, ThermoFisher Scientific). RT-qPCR of the selected markers (Crybb1-Mm00517828_m1; Ebf3-Mm00438642_m1; Garnl3-Mm00724806_m1; Gpr34-Mm02620221_s1; Lag3-Mm00493071_m1; Nuak1-Mm01250704_m1; Olfml3-Mm00513567_m1; Rtn1-Mm00463222_m1; Rtn4rl1-Mm00710554_m1; Sall1-Mm00491266_m1; Sall3-Mm00774693_g1; Siglech-Mm00618627_m1; Slc1a3-Mm00600697_m1; Sparc-Mm00486332_m1; Tnfrsf17-Mm00495683_m1) was simultaneously performed by the means of a Custom TaqMan Array Card (Applied Biosystems by Life Technologies) using Vii7 (ViiA 7 Real-Time PCR System, Applied Biosystems). Results were normalized according to the mean value of the following housekeeping genes (Atf3-Mm00476032_m1, Gusb-Mm01197698_m1, Hprt-Mm01545399_m1, Mertk-Mm00434920_m1, Spint1-Mm00444186_m1) and analyzed using the RQ manager v1.2.1 and the DataAssistv3.0 software (Applied Biosystems).
Targeted flow cytometry validation.
EAE was induced in wild-type mice as previously described and mice were killed at pre-onset (8–9 dpi), at the peak of disease (16–18 dpi), and in the chronic phase (38–40 dpi). Microglia and CNS leukocytes were isolated and labeled with fluorochrome-conjugated mAbs specific for mouse CD11b (clone M1/70), CD45 (clone 30-F11), Ly6C (clone AL-21; all purchased from BD Biosciences), F4/80 (clone BM8), Siglech (clone 551), Lag-3 (clone C9BTW), and appropriate isotype controls (BioLegend), Tnfsfr17 (BCMA, clone 161616) and its appropriate isotype control (R&D Systems). Dead cells were always excluded using a LIVE/DEAD stain kit (Invitrogen). Flow cytometry was performed using CantoII flow cytometer (Becton Dickinson) and analyzed with FlowJo software (Tree Star).
Experimental design and statistical analyses.
The primary aim of this study was to identify specific microglia markers throughout life and upon inflammation. For the preparation of E14, P1 P60 microglia and E14, P1 liver myeloid samples, cells were obtained as previously described. Genome-wide transcription study has been made on three to four biological replicates of microglia and liver myeloid cells at each time point. Each replicate in turn derived from a pool of 10 mice each. Statistical details for microarray analyses can be found in the genome-wide expression analysis paragraph.
Transcriptomics datasets generated in this study were uploaded in ArrayExpress database (E-MTAB-8059), whereas the transcriptomics dataset relative to unstimulated, M1 and M2 polarized mouse macrophages was described by Garzetti et al., 2014 and Colombo et al. (2018) and recovered from ArrayExpress database (accession no. E-MTAB-6416).
Microglia and monocyte-derived macrophage (MoDM) samples from EAE mice, including chimeric mice, were obtained as previously described. Each experimental group consisted of two independent biological replicates derived from the pool of two samples. Each sample was derived from a pool of 10 brain and spinal cords. Quantitative PCR data were performed on a Custom TaqMan Array Card (Applied Biosystems by Life Technologies) using Vii7 instrument (ViiA 7 Real-Time PCR System, Applied Biosystems). Results were normalized according to the mean value of the following housekeeping genes (Atf3-Mm00476032_m1, Gusb-Mm01197698_m1, Hprt-Mm01545399_m1, Mertk-Mm00434920_m1, Spint1-Mm00444186_m1) and analyzed using the RQ manager v1.2.1 and the DataAssistv3.0 software (Applied Biosystems).
For the flow cytometry validation experiments, EAE was induced in wild-type mice as previously described and mice were killed at pre-onset (8–9 dpi), at the peak of disease (16–18 dpi), and in the chronic phase (38–40 dpi). Microglia and CNS leukocytes were isolated land labeled with the appropriate mAbs, as previously described. Dead cells were always excluded using a LIVE/DEAD stain kit (Invitrogen). Flow cytometry was performed using Canto II flow cytometer (Becton Dickinson) and analyzed with FlowJo software (Tree Star). Each sample derived from a pool of 5–7 mice. Statistical analysis was performed using Prism 6 software (GraphPad Software; RRID:SCR_002798). For statistical tests between groups, unpaired t test was used. The threshold for significance for all experiments was set at *p < 0.05. Smaller p values were represented as **p < 0.01 and ***p < 0.001. In bar graphs, all data are plotted as mean ± SEM.
Results
Transcriptional profiling of CNS and liver myeloid cells during development
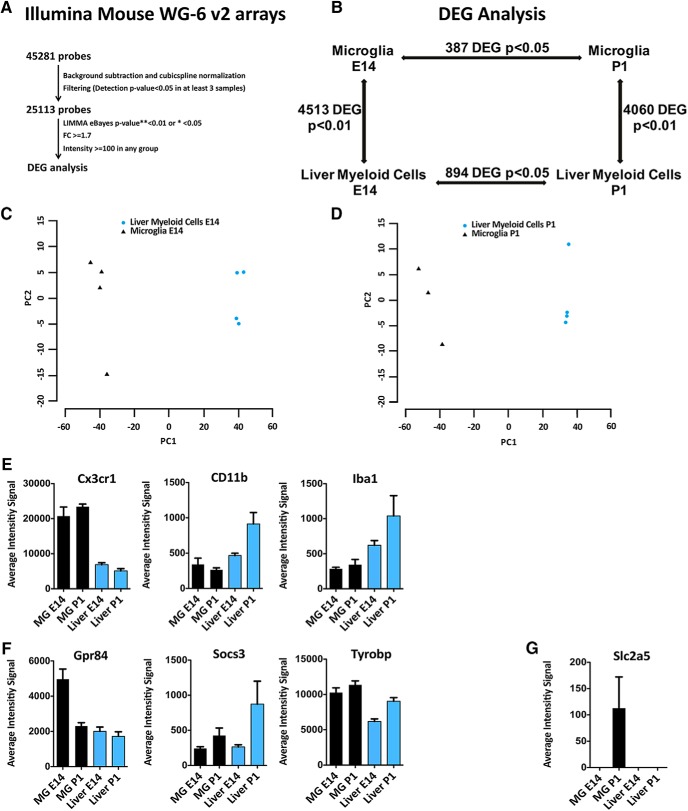
To study the transcriptional profiles of peripheral and central myeloid cells during development, we isolated microglia and liver macrophages at E14 and P1. Indeed, after blood–brain barrier closing at E14.5, CNS myeloid cells remain isolated from further influx of myeloid cells, whereas fetal liver macrophages generate all other tissue-resident macrophages and monocytes derived macrophages (MoDMs) until the second postnatal week when bone-marrow hematopoiesis starts (Hoeffel and Ginhoux, 2018; Li and Barres, 2018). CD11b+ CD45low microglia and CD11b+ CD45high liver macrophages were sorted from naive C57BL6 E14 embryos and P1 pups, RNA was extracted and analyzed by gene expression microarrays. Briefly, raw data were subjected to background subtraction followed by cubic spline normalization. Filtering was performed for probes with detection p < 0.05 in at least three samples. The significance threshold was set to corrected p < 0.05 for the intra-tissue comparison and corrected p < 0.01 in case of inter-tissue comparison and the DEGs were defined by statistical significance and thresholds on fold-change (±1.7) and expression intensity (>100 in any 1 of the 2 compared groups; Fig. 1A).
Figure 1.
Transcriptional profiling of microglia and liver myeloid cells during development. Genome-wide expression analysis of microglia and liver macrophages sorted from naive C57BL6 E14 embryos and P1 pups. A, Raw data were subjected to background subtraction followed by cubic spline normalization. Filtering was performed for probes with detection p < 0.05 in at least three samples. The significance threshold was set to p < 0.05 for the intra-tissue comparison and p < 0.01 for inter-tissue comparison. Differentially expressed genes (DEGs) were defined by statistical significance and thresholds on fold-change (±1.7) and expression intensity (>100 in any 1 of the 2 compared groups). B, DEGs (4513 and 4060) were found comparing microglia and liver myeloid cells at E14 and P1, respectively, whereas only 387 and 894 DEGs were found between embryonic and postnatal microglia or liver myeloid cells, despite the less stringent statistical threshold (corrected p < 0.05). C, D, PCA plots of microglia versus liver macrophages at E14 (C) and P1 (D). E, Simultaneous expression of pan myeloid genes in microglia and liver myeloid cells at E14 and P1. F, Referred microglial-specific markers were expressed by both microglia and liver myeloid cells at E14 and P1. G, A well known microglia-specific marker (i.e., Slc2a5) was absent in liver myeloid cells at both time points but undetectable in microglia at E14 and barely expressed at P1. Each bar represents the average intensity signal ± SEM of three biological replicates per time point, resulting from a pool of 10 mice each.
We found 4513 and 4060 DEGs passing p < 0.01 when comparing brain and liver myeloid cells at E14 and P1 respectively, indicating extensive differences between myeloid cells in the two distinct compartments, at both time points. The comparison between myeloid cells at E14 and P1 in the same compartment on the contrary evidenced only 387 and 894 DEGs between embryonic and postnatal microglia or liver myeloid cells respectively despite the application of a less stringent statistical threshold (p < 0.05), thus highlighting minor transcriptional changes in each compartment between E14 and P1 (Fig. 1B–D). We can conclude that in the analyzed experimental conditions tissue-related myeloid transcriptional reprogramming is more extensive than time-related changes. Common myeloid genes, such as Cx3cr1, CD11b, Iba1 were expressed by microglia and liver myeloid cells even if at different levels (Fig. 1E). Notably, the well known microglia-specific markers Gpr84, Socs3, and Tyrobp (Gautier et al., 2012; Chiu et al., 2013; Hickman et al., 2013; Butovsky et al., 2014; Zhang et al., 2014; Crotti and Ransohoff, 2016; Wolf et al., 2017), were expressed by liver myeloid cells at comparable levels (Fig. 1F), whereas Slc2a5 was undetectable in microglia at E14 and barely detectable at P1 (Fig. 1G).
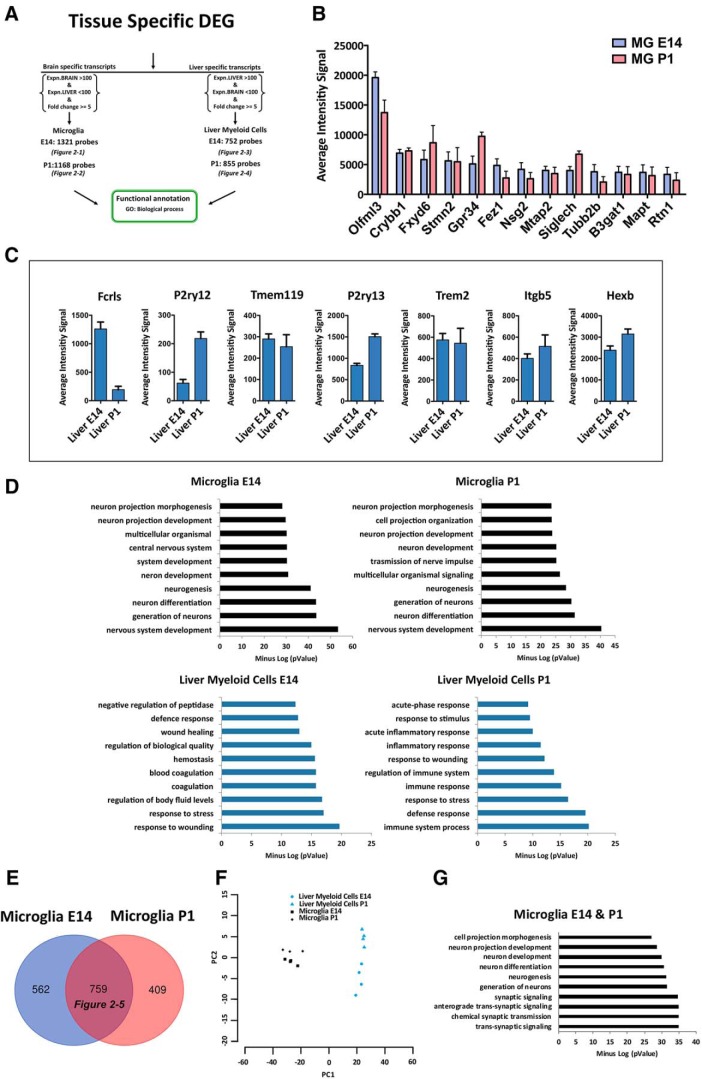
To identify transcripts exclusively expressed by microglia or liver myeloid cells at each time point, among the tissue-related transcriptional profiles we selected those probes with no expression in the other tissue and a fold-change ≥ 5. We ended up with an E14 microglia-specific list consisting of 1321 DEGs, a P1 microglia-specific list (1168 DEGs), an E14 liver-specific list (752 DEGs), and finally a P1 liver-specific list (855 DEGs; Fig. 2A; Fig. 2-1; Fig. 2-2; Fig. 2-3; Fig. 2-4). Interestingly, among the top expressed genes the E14 and P1 microglia-specific lists contained well known microglia markers such as Olfml3, Crybb1, Gpr34, Siglech (Gautier et al., 2012; Chiu et al., 2013; Hickman et al., 2013; Butovsky et al., 2014; Zhang et al., 2014; Crotti and Ransohoff, 2016; Wolf et al., 2017) and also transcripts known to be highly expressed by neurons and oligodendrocyte precursors, such as Fxyd6, Stmn2, Fez1, Nsg2, Mtap2, Tubb2, B3gat1, Mapt, Rtn1 (Zhang et al., 2014; Fig. 2B; Fig. 2-1; Fig. 2-2). Of note, some widely referred microglia markers such as Fcrls, P2ry12, Tmem119, P2ry13, Trem2, Itgb5, Hexb (Gautier et al., 2012; Chiu et al., 2013; Hickman et al., 2013; Butovsky et al., 2014; Zhang et al., 2014; Crotti and Ransohoff, 2016; Wolf et al., 2017) were excluded from both microglia-specific lists because they were expressed also in liver myeloid cells (Fig. 2C).
Figure 2.
Identification of microglia-specific transcripts during development. A, Identification of tissue-specific transcripts [1321 E14 microglia (Figure 2-1); 1168 P1 microglia (Figure 2-2); 752 E14 liver myeloid cells (Figure 2-3); and 855 P1 liver myeloid cells (Figure 2-4)] expressed by microglia or liver myeloid cells at each time point with no expression in the other tissue and a fold-change ≥ 5. B, Top expressed transcripts in microglia-specific lists at E14 and P1. C, Well known microglia-specific markers were also detectable in liver myeloid cells and therefore eliminated from further analysis. Each bar represents the average intensity signal ± SEM of three biological replicates per time point, resulting from a pool of 10 mice each. D, Top significant biological processes enriched in each tissue-specific myeloid signature at E14 and P1. E, F, Identification of a core of 759 conserved transcripts, which are microglia-specific during development (Figure 2-5) and relative PCA plot (F). G, Top significant biological processes enriched in microglia-specific lists at E14 and P1.
Microglia E14-Specific Transcripts. List of 1321 transcripts exclusively expressed by Microglia at E14 (with no expression in Liver myeloid cells at E14 and a fold change equal to or higher than 5). Download Figure 2-1, XLSX file (112.2KB, xlsx)
Microglia P1-Specific Transcripts. List of 1168 transcripts exclusively expressed by Microglia at P1 (with no expression in Liver myeloid cells at P1 and a fold change equal to or higher than 5). Download Figure 2-2, XLSX file (99.1KB, xlsx)
E14 Liver Myeloid Cells-Specific Transcripts. List of 752 transcripts exclusively expressed by Liver myeloid cells at E14 (with no expression in Microglia at E14 and a fold change equal to or higher than 5). Download Figure 2-3, XLSX file (68.8KB, xlsx)
P1 Liver Myeloid Cells-Specific Transcripts. List of 855 transcripts exclusively expressed by Liver myeloid cells at P1 (with no expression in Microglia at P1 and a fold change equal to or higher than 5). Download Figure 2-4, XLSX file (76.7KB, xlsx)
Microglia Specific signatures during embryonic and early postnatal life. List of 759 Microglia Specific Transcripts both at E14 and P1. Download Figure 2-6, XLS file (130.5KB, xls)
Bioinformatic analyses of the biological processes enriched in each tissue-specific list unraveled that the top significant functions were related to neurogenesis and neural development in the E14 and P1 microglia lists and to immune response in the liver myeloid cell lists (Fig. 2D), indicating distinct potential functional properties of the myeloid cells in the two compartments during development.
Finally, by crossing E14 and P1 microglia-specific signatures we selected a novel core of 759 transcripts, which represent conserved microglia-specific markers during embryonic and early postnatal life (Fig. 2E,F; Fig. 2-5). Top gene ontology terms in this signature were again related to neurogenesis and neural development (Fig. 2G).
Transcriptional profiling of adult microglia under resting and pathological conditions
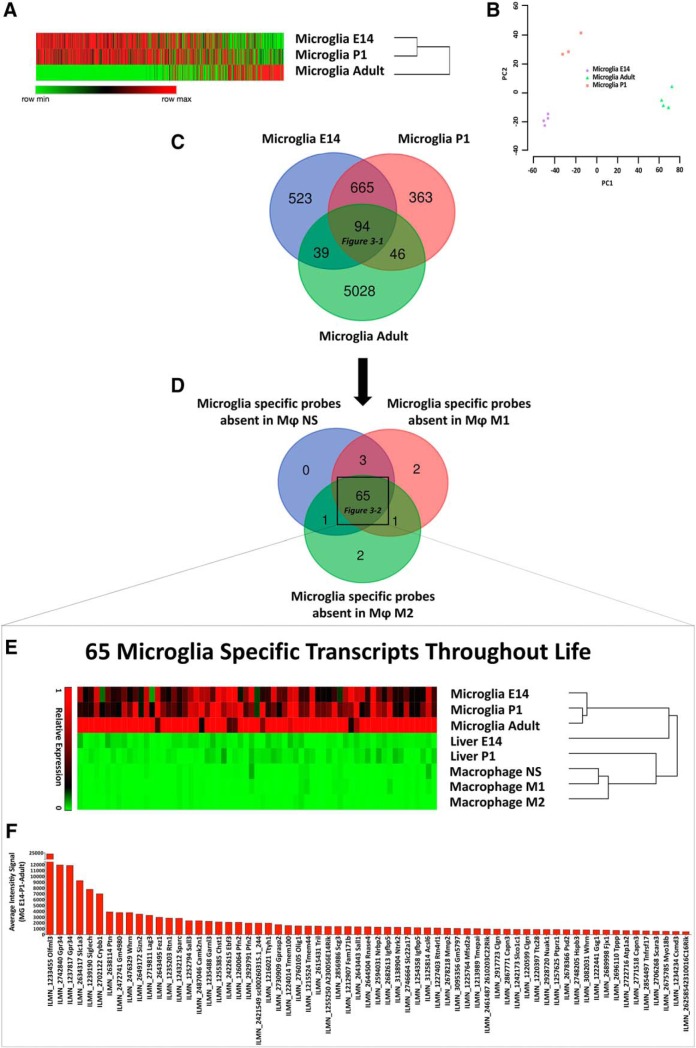
To assess to what extent the transcriptional profile of adult brain microglia differs from the one found during development, we analyzed in house-generated transcriptomes of adult microglia sorted from the CNS of naive C57BL/6 mice, and found 3353 and 1876 DEGs passing corrected p < 0.01 when adult microglia were compared with E14 and P1 microglia, respectively. This observation demonstrates that microglia undergo extensive transcriptional reprogramming from the embryonic/postnatal stage to the adult state (Fig. 3A,B).
Figure 3.
Identification of 65 homeostatic microglia-specific transcripts throughout life. A, B, Heatmap and hierarchical clustering of differentially expressed genes in microglia at E14, P1, and in adulthood. Each lane represents the average expression value of three biological replicates per time-point; relative PCA plot (B). C, Venn diagram displays unique and intersecting DEGs among microglia at E14, P1, and in adulthood. Ninety-four transcripts were shared at all time points (Figure 3-1). D, Venn diagram displaying homeostatic microglia-specific transcripts with no expression in adult peritoneal macrophages at steady state or after differentiation toward an M1 or M2 phenotype. Sixty-five transcripts were absent in all three macrophage populations (Figure 3-2). E, Heatmap and hierarchical clustering of the identified 65 microglia-specific transcripts in microglia at E14, P1, in adulthood, and in liver macrophages at E14 and P1, and peritoneal macrophages at steady state or after differentiation toward an M1 or M2 phenotype. Each lane represents the average expression value of three biological replicates per time point, in turn deriving from polls of 8–10 mice each. F, Average gene expression of the identified 65 microglia-specific transcripts at E14, P1, and adult phase.
List of 94 Microglia conserved transcripts not present in liver myeloid cells but always expressed by microglia at E14, P1 and in adulthood. Download Figure 3-1, XLS file (35.5KB, xls)
65 Microglia Specific Transcripts Throughout Life. List of 65 homeostatic microglia-specific transcripts, which are always expressed by microglia throughout life at E14, P1and in adulthood, but neither expressed in liver macrophages at E14 and P1 nor in adult peritoneal macrophages at steady state or after differentiation towards an M1 or M2 phenotype. Download Figure 3-2, XLSX file (22KB, xlsx)
However, there was a core of 94 conserved transcripts not present in liver myeloid cells and expressed by microglia at E14, P1, and in the adult stage (Fig. 3B; Fig. 3-1).
To exclude transcripts present in adult macrophages either in resting condition or upon stimulation, we recovered transcriptomes relative to unstimulated peritoneal macrophages and M1 or M2 differentiated macrophages from our published study (Garzetti et al., 2014; Colombo et al., 2018) and excluded all those genes among the 94 selected microglia-specific transcripts that were expressed by macrophages in any condition. We obtained a final list of 65 homeostatic microglia-specific transcripts, which were always present in microglia throughout life at E14, P1and in adulthood, but not expressed in liver macrophages at E14 and P1 or adult peritoneal macrophages at steady state or after differentiation toward and M1 or M2 phenotype (Fig. 3D–F; Fig. 3-2).
Literature mining indicated that among the 65 microglia-specific probes, 14 genes (Crybb1, Garnl3, Gpr34, Lag3, Nuak1, Olfml3, Rtn1, Rtn4rl1, Sall1, Sall3, Siglech, Slc1a3, Sparc, Tnfrsf17) appeared also as microglia-specific in other published studies (Gautier et al., 2012; Chiu et al., 2013; Hickman et al., 2013; Butovsky et al., 2014). Thus, we further tested the specificity of the 14 markers searching for their expression in MoDMs and microglia during the whole course of EAE.
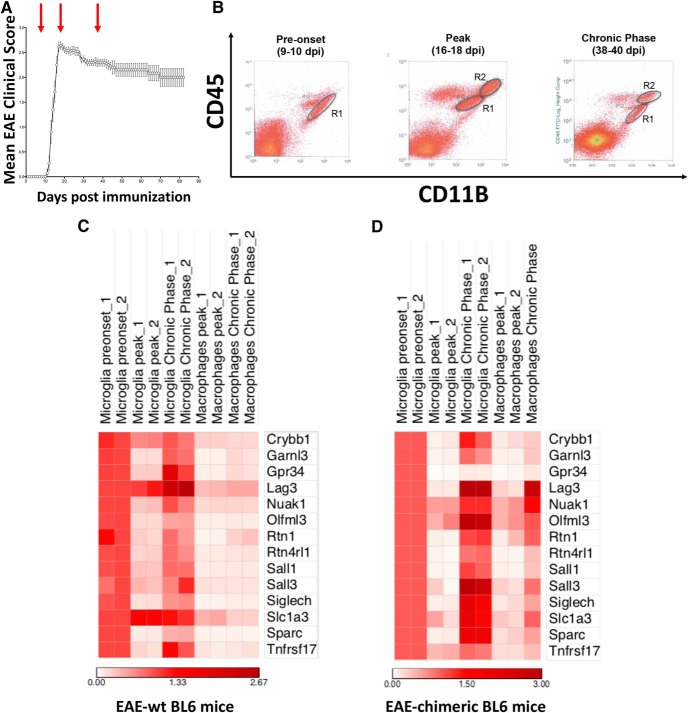
For that purpose, we induced EAE in C57BL6 female mice and sorted out CD11b+ CD45low microglia and CD11b+ CD45high MoDMs from brain and spinal cord at pre-onset (9–10 dpi), peak (14–16 dpi), and chronic phase (38–40 dpi) of the disease (Fig. 4A,B). RNA was extracted, retro-transcribed, and the levels of the abovementioned 14 genes were measured by a custom-made TaqMan card. As shown in Figure 4C, they showed an overall transient downregulation in microglia in the acute phase of EAE, because their levels were lower at the peak of disease compared with the pre-onset phase but substantially regained their basal expression in the chronic phase. Notably, these transcripts were detected in MoDMs, although at much lower levels than in microglia cells. To better discriminate between microglia and MoDMs in inflamed CNS, we generated bone marrow chimeric mice reconstituted with cells expressing the GFP reporter and induced EAE in such mice. Gene expression analysis performed on sorted resident microglia (GFP− CD11b+ CD45low) and MoDMs (GFP+ CD11b+ CD45high) confirmed what observed in wild-type mice. In fact, all the 14 putative microglia-specific markers were also expressed by MoDMs, barely at the peak of disease but with concomitant upregulation in the chronic phase, which in some cases (i.e., Lag3, Nuak1, Olfml3, Rtn1, Sall3, Slc1a3) reached comparable expression levels as in microglia (Fig. 4D).
Figure 4.
MoDMs express microglia-specific markers during EAE. A, EAE induction and expression. Red arrows indicate time points used for isolation of CNS myeloid cells (pre-onset, peak, and chronic phase). B, Exemplificative plots of the sorted populations at each time point: R1 (microglia), R2 (MoDMs). C, D, Heatmap reporting mRNA levels of 14 selected microglia-specific genes in sorted microglia and MoDMs during EAE in wild-type mice (C) and chimeric mice (D). Data represent two independent experiments and each sample results from a pool of 10 mice.
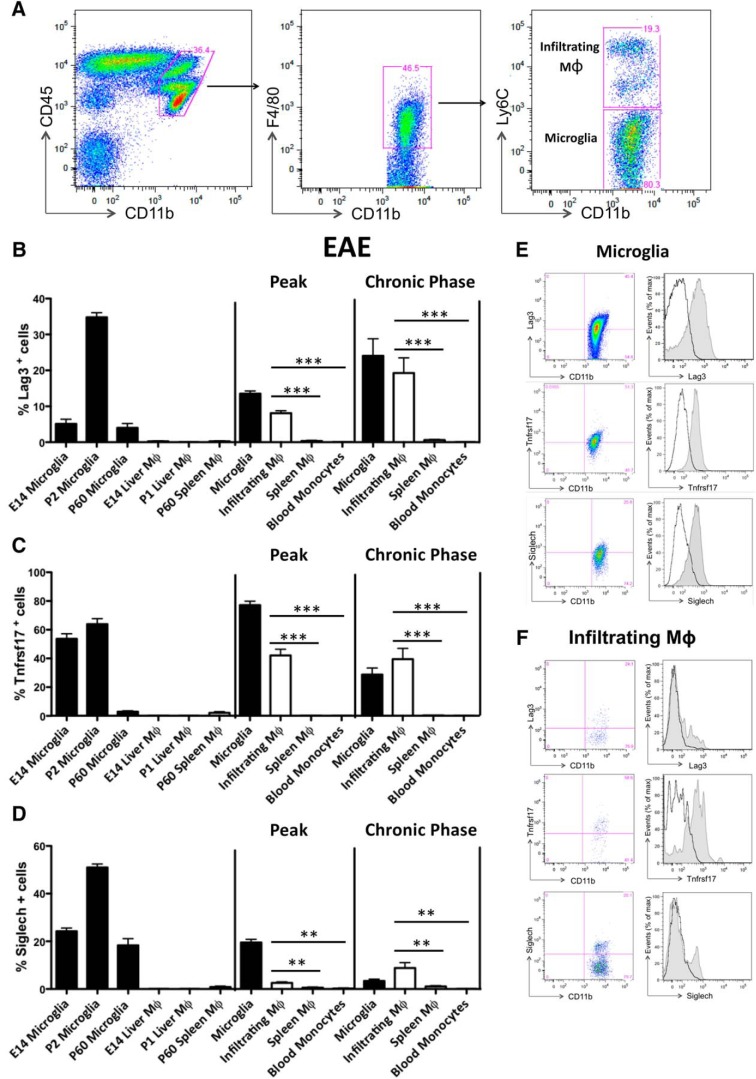
Finally, to evaluate protein expression of putative microglia-specific markers, we selected Lag3, Tnfrsf17, and Siglech because of their localization on cell membrane and the availability of specific flow cytometry antibodies and measured their protein levels on myeloid cells during development and in adulthood under physiological and pathological conditions. We found that they were all highly expressed by microglia at E14, P1 and, to a lesser extent, in the adult phase. In accordance with our transcriptomics data, they were absent on cell membrane of liver macrophages during development, at E14 and P1, and of adult splenic macrophages under physiological conditions. Notably, in the acute phase of EAE these markers were maintained by microglia but acquired by CNS infiltrating MoDMs as well, whereas not expressed by spleen or blood myeloid cells. Similar observations were obtained for the chronic phase of EAE [Fig. 5A–F; (B) Peak, infiltrating MoDMs vs spleen macrophages: t = 11.45, df = 10, p < 0.0001; infiltrating MoDMs vs blood monocytes: t = 12.07, df = 10, p < 0.0001; Chronic phase, infiltrating MoDMs vs spleen macrophages: t = 3.932, df = 27, p = 0.0005; infiltrating MoDMs vs blood monocytes: t = 3.834, df = 26, p = 0.0007; (C) Peak, infiltrating MoDMs vs spleen macrophages: t = 6.692, df = 16, p < 0.0001; infiltrating MoDMs vs blood monocytes: t = 6.715, df = 16, p < 0.0001; Chronic phase, infiltrating MoDMs vs spleen macrophages: t = 5.701, df = 20, p < 0.0001; infiltrating MoDMs vs blood monocytes: t = 5.203, df = 18, p < 0.0001; (D) Peak, infiltrating MoDMs vs spleen macrophages: t = 3.165, df = 16, p = 0.0060; infiltrating MoDMs vs blood monocytes: t = 3.759, df = 16, p = 0.0017; Chronic phase, infiltrating MoDMs vs spleen macrophages: t = 3.298, df = 30; p = 0.0025; infiltrating MoDMs vs blood monocytes: t = 3.210, df = 27, p = 0.0034]. All together these data indicate that such markers are truly microglia-specific under physiological conditions but not during CNS inflammation as acquired specifically by infiltrating MoDMs.
Figure 5.
Microglia-specific markers are acquired specifically by peripheral myeloid cells entering the CNS. A, Flow cytometry of CNS leukocytes from EAE mice, assessed at the chronic phase (38–40 dpi). Frequencies of microglia (CD11b+ CD45low F4/80+ Ly6Clow cells) and MoDMs (CD11b+ CD45high F4/80+ Ly6Chigh cells) are shown in the plots. B–D, Graphs show the frequency of Lag3+, Tnfrsf17+, and Siglech+ on microglia at E14, P1, P60, liver macrophages at E14 and P1, adult splenic macrophages in physiological conditions as well as on microglia, MoDMs, spleen macrophages, and blood monocytes in the acute phase and chronic phases of EAE (n = 5 mice per group). Data represent the mean ± SEM and are representative of two independent experiments. ***p ≤ 0.001, **p ≤ 0.01, unpaired t test. E, F, FACS analysis of Lag3+, Tnfrsf17+, and Siglech+ microglia (E) and MoDMs (F) at the chronic phase of EAE. Black lines represent isotype controls. Data are representative of three or more replicates.
Discussion
Here we provide evidence that the phenotypic differentiation between microglia and peripheral macrophages is age- and context-dependent, so that peripheral macrophages may express some of the most commonly described microglia markers in the literature (Gautier et al., 2012; Chiu et al., 2013; Hickman et al., 2013; Butovsky et al., 2014; Zhang et al., 2014). This discrepancy can be ascribed to developmental stages, distinct myeloid cell populations under analysis and, not last, healthy rather than pathological conditions tested.
The first clue comes from the transcriptional comparison between embryonic (E14.5) and early postnatal (P1) microglia with liver macrophages at the same time points. We found large transcriptional changes between myeloid cells in the two distinct compartments (brain and liver), and only minor time-related intra-compartment changes, thus confirming that the microenvironment is the major force driving tissue-resident macrophages transcriptional reprogramming already from early development (Mass et al., 2016; Bennett et al., 2018). Interestingly, our data also unraveled that liver macrophages can express known microglia markers, such as Gpr84, Socs3, and Tyrobp (Crotti and Ransohoff, 2016; Wolf et al., 2017). This finding is original and relevant. It is original because the available transcriptome studies have described either adult microglia and macrophages in different settings (Beutner et al., 2013; Crotti and Ransohoff, 2016) or microglia during development without any comparison with peripheral myeloid cells (Matcovitch-Natan et al., 2016; Hammond et al., 2019; Masuda et al., 2019). It is relevant because it shows that some phenotypic convergence between the two cell types may occur during development, despite their distinct ontogeny (Hoeffel and Ginhoux, 2018; Li and Barres, 2018).
Next, among the tissue-related transcriptional profiles, we selected those probes with no expression in the other tissue, obtaining four myeloid signatures (E14 microglia-specific list, P1 microglia-specific list, E14 liver-specific list, and P1 liver-specific list). Significantly enriched processes in microglia-specific signatures were related to neurogenesis and neural development and not to immune function as they were in liver macrophages. Moreover, within the top E14 and P1 microglia-specific transcripts, in addition to well known microglia-specific markers such as Olfml3, Crybb1, Gpr34, Siglech (Crotti and Ransohoff, 2016; Wolf et al., 2017) we found the presence of genes known to be highly expressed by neurons and oligodendrocytes precursors such as Fxyd6, Stmn2, Fez1, Nsg2, Mtap2, Tubb2, B3gat1, Mapt, Rtn1 (Zhang et al., 2014). Of note, some widely referred microglia-specific markers such as Fcrls, P2ry12, Tmem119, P2ry13, Trem2, Itgb5, Hexb were excluded from both microglia-specific lists because they were also detectable, even though barely, in liver myeloid cells.
Finally, by crossing E14 and P1 microglia-specific signatures we selected a novel core of 759 transcripts, which represent conserved microglia-specific markers during embryonic and early postnatal life. Altogether, our study provides a novel source of microglia-specific markers during embryonic and early postnatal life, useful for the generation of hypothesis-driven experiments about microglia function, especially in relation to neurogenesis and brain development.
The addition of data from the transcriptional profiling of adult cells shows a large time-related transcriptional reprogramming in adult microglia compared with embryonic/postnatal stages, possibly reflecting the different functions exerted by microglia throughout life, which are mainly related to neurogenesis, synapsis maturation, and myelin formation during development (Schafer and Stevens, 2015) and to brain homeostasis later on (Salter and Stevens, 2017; Wolf et al., 2017). Nonetheless, we identified a core of 65 homeostatic microglia-specific transcripts, always present in microglia throughout life and not expressed in liver E14 and P1 macrophages or in adult peritoneal macrophages at steady state or after differentiation toward the M1 or M2 phenotypes. Such a core of stable microglia-specific genes has not been defined by any previously published transcriptome studies (Crotti and Ransohoff, 2016), where the definition of specificity for microglia is simply based on the enrichment in transcripts and not by the systematic absence of expression in peripheral myeloid cells. The latter indeed, has been experimentally confirmed only in adult macrophages for a few markers, such as P2ry12 (Haynes et al., 2006), Fcrls (Butovsky et al., 2014), Tmem119 (Bennett et al., 2016), Sall1 (Buttgereit et al., 2016; Koso et al., 2016), and Siglech (Konishi et al., 2017). On the other hand, the more recent generation of bulk and single-cell RNA-sequencing data highlights an ever-growing (and at times contradictory) complexity of microglia phenotypes (Hagemeyer et al., 2017; Wlodarczyk et al., 2017; Hammond et al., 2019; Masuda et al., 2019), which may result from variables as brain region, ontogeny, disease, age, sex, experimental procedures, and species (Dubbelaar et al., 2018). Despite this possible heterogeneity, our list of 65 transcripts, which are microglia-specific throughout life and absent in peripheral macrophages, represents a relevant source for the development of more appropriate transgenic mice for microglia targeting.
Yet, the main evidence of this study comes from the validation experiment of the microglia-specific markers under pathological conditions, as it demonstrates their expression by MoDMs during EAE. This was unexpected, as we have selected a subset of transcripts (Crybb1, Garnl3, Gpr34, Lag3, Nuak1, Olfml3, Rtn1, Rtn4rl1, Sall1, Sall3, Siglech, Slc1a3, Sparc, Tnfrsf17) that appeared microglia-specific in our datasets and in the literature (Crotti and Ransohoff, 2016). To exclude the misclassification of peripheral versus central myeloid cells, we repeated the experiment using the GFP-bone marrow chimeric. Altogether we found that all the markers were strongly downregulated in microglia in the acute phase of EAE but regained their basal level in the chronic phase. This is in line with previous studies showing the drastic downregulation of homeostatic microglia markers toward a proinflammatory signature during the acute phase of EAE (Butovsky et al., 2014; Buttgereit et al., 2016; Mrdjen et al., 2018; Jordão et al., 2019). Notably, the same observation is confirmed in active multiple sclerosis lesions (Zrzavy et al., 2017; Jordão et al., 2019). Most importantly, these markers had some expression also in MoDMs, where they were barely detectable in acute EAE but enhanced in the chronic phase, with levels in some cases comparable to those reached in microglia (i.e., Lag3, Nuak1, Olfml3, Rtn1, Sall3, Slc1a3).
Finally, we searched for protein validation of three membrane receptors (i.e., Lag3, Tnfrsf17, and Siglech), and confirmed by flow cytometry their selective expression by microglia throughout life under physiological conditions and their appearance on MoDMs only along the course of EAE. Interestingly, we observed that their expression was strictly confined to CNS infiltrating macrophages because blood monocytes and spleen macrophages did not upregulate these markers along the entire course of the disease. This finding challenges the definition of microglia-specific markers based on recent transcriptomic studies (Crotti and Ransohoff, 2016; Wolf et al., 2017). This can be explained because, as already mentioned, the term “microglia-specific” has mainly been used as synonymous with “highly enriched”. Second, a couple of transcriptomic studies describe a myeloid cell phenotype during EAE and conclude that microglia and MoDMs maintain their own distinct expression profile (Butovsky et al., 2014; Buttgereit et al., 2016; Jordão et al., 2019), but these analyses were just limited to the acute and not the chronic phase of the disease. MoDMs convergence toward a “microglia phenotype” in our model is supported by the finding of Sall1 downregulation in microglia in acute EAE followed by its increased expression in MoDMs in chronic EAE. Indeed, the transcription factor Sall1 was recently described as crucial to maintain microglial identity in the CNS environment, because microglia-specific deficiency of Sall1 leads to rapid loss of microglial signature with concomitant impaired neurogenesis and upregulation of genes associated with inflammatory macrophages (Buttgereit et al., 2016; Gosselin et al., 2017). Our observation that Sall1 expression is present in monocyte-derived macrophages especially during the chronic phase of EAE supports the hypothesis that the fraction of macrophages expressing microglial markers may have an anti-inflammatory, protective role. Overall, findings showing on the one hand acquisition of a microglia-like phenotype by MoDMs in the chronic phase of EAE (as in our study) and on the other hand microglia shift to a proinflammatory macrophage-like phenotype in the acute phase (Lewis et al., 2014; Buttgereit et al., 2016; Zrzavy et al., 2017; Mrdjen et al., 2018; Jordão et al., 2019) suggest the existence of bidirectional plasticity, according to which in the acute phase of EAE microglia acquire a detrimental “proinflammatory macrophage” phenotype, whereas in the chronic phase MoDMs acquire a microglia phenotype, which is essential for full recovery. The longer time course of our expression study stretched until the chronic phase of EAE, the first in this regard to our knowledge, underlines how time is crucial for macrophage plasticity to become evident. Indeed, MoDMs, long known as highly plastic cells (Mantovani et al., 2013), has been recently shown to differentiate into specialized tissue-resident macrophages, not only in the lung and liver (Scott et al., 2016; van de Laar et al., 2016) but also into microglia under certain circumstances, when their niche is vacant after genetic or pharmacological ablation (Guilliams and Scott, 2017). Microglia themselves result to be highly dependent on their environment, because they quickly lose their specific signatures when cultured but regain it once transferred back to the brain (Bohlen et al., 2017; Gosselin et al., 2017).
Overall, the focus of our study was to identify stable microglial markers throughout life and validate a selection of them during neuroinflammation. Whether microglia and macrophages do bear distinctive markers at different stages of EAE remains an open relevant issue, which deserves future investigation.
In conclusion, we speculate that myeloid cell reprogramming is time- and context-dependent and thus the general definition of microglia-specific markers may result incorrect, particularly under pathological conditions. Further, our data suggest that in monophasic self-limiting CNS inflammatory conditions microglia and MoDMs may converge phenotypically and act as a functional unit rather than as different entities, while (partly) acquiring their respective original signatures to boost their mutual functions in different phases of the disease. Whether or not this functional synergism can apply also to other pathological contexts and whether chronic diseases occur when this synergic system fails needs further investigation.
Footnotes
This work was supported in part by Merk-Serono, TargetBrain (EU Framework 7 project HEALTH-F2–2012-279017) and Fondazione Italiana Sclerosi Multipla (FISM). We thank Elena Brambilla and Annamaria Finardi, San Raffaele Scientific Institute, for technical assistance; Dejan Lazarevic, Center for Translational Genomics and BioInformatics, San Raffaele Scientific Institute, for technical assistance; Flow cytometry Resource, Advanced Cytometry Technical Applications Laboratory (FRACTAL Core Facility), San Raffaele Scientific Institute, for technical help and FACS sorting.
The authors declare no competing financial interests.
References
- Antony JM, Paquin A, Nutt SL, Kaplan DR, Miller FD (2011) Endogenous microglia regulate development of embryonic cortical precursor cells. J Neurosci Res 89:286–298. 10.1002/jnr.22533 [DOI] [PubMed] [Google Scholar]
- Arnò B, Grassivaro F, Rossi C, Bergamaschi A, Castiglioni V, Furlan R, Greter M, Favaro R, Comi G, Becher B, Martino G, Muzio L (2014) Neural progenitor cells orchestrate microglia migration and positioning into the developing cortex. Nat Commun 5:5611. 10.1038/ncomms6611 [DOI] [PubMed] [Google Scholar]
- Bennett FC, Bennett ML, Yaqoob F, Mulinyawe SB, Grant GA, Hayden Gephart M, Plowey ED, Barres BA (2018) A combination of ontogeny and CNS environment establishes microglial identity. Neuron 98:1170–1183.e8. 10.1016/j.neuron.2018.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A, Weissman IL, Chang EF, Li G, Grant GA, Hayden Gephart MG, Barres BA (2016) New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A 113:E1738–1746. 10.1073/pnas.1525528113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutner C, Linnartz-Gerlach B, Schmidt SV, Beyer M, Mallmann MR, Staratschek-Jox A, Schultze JL, Neumann H (2013) Unique transcriptome signature of mouse microglia. Glia 61:1429–1442. 10.1002/glia.22524 [DOI] [PubMed] [Google Scholar]
- Bohlen CJ, Bennett FC, Tucker AF, Collins HY, Mulinyawe SB, Barres BA (2017) Diverse requirements for microglial survival, specification, and function revealed by defined-medium cultures. Neuron 94:759–773.e8. 10.1016/j.neuron.2017.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu L, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, Weiner HL (2014) Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci 17:131–143. 10.1038/nn.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit A, Lelios I, Yu X, Vrohlings M, Krakoski NR, Gautier EL, Nishinakamura R, Becher B, Greter M (2016) Sall1 is a transcriptional regulator defining microglia identity and function. Nat Immunol 17:1397–1406. 10.1038/ni.3585 [DOI] [PubMed] [Google Scholar]
- Cardona AE, Huang D, Sasse ME, Ransohoff RM (2006) Isolation of murine microglial cells for RNA analysis or flow cytometry. Nat Protoc 1:1947–1951. 10.1038/nprot.2006.327 [DOI] [PubMed] [Google Scholar]
- Chiu IM, Morimoto ET, Goodarzi H, Liao JT, O'Keeffe S, Phatnani HP, Muratet M, Carroll MC, Levy S, Tavazoie S, Myers RM, Maniatis T (2013) A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep 4:385–401. 10.1016/j.celrep.2013.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo F, Bastoni M, Nigro A, Podini P, Finardi A, Casella G, Ramesh M, Farina C, Verderio C, Furlan R (2018) Cytokines stimulate the release of microvesicles from myeloid cells independently from the P2X7 receptor/acid sphingomyelinase pathway. Front Immunol 9:204. 10.3389/fimmu.2018.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti A, Ransohoff RM (2016) Microglial physiology and pathophysiology: insights from genome-wide transcriptional profiling. Immunity 44:505–515. 10.1016/j.immuni.2016.02.013 [DOI] [PubMed] [Google Scholar]
- Cunningham CL, Martínez-Cerdeño V, Noctor SC (2013) Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J Neurosci 33:4216–4233. 10.1523/JNEUROSCI.3441-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derecki NC, Cronk JC, Lu Z, Xu E, Abbott SB, Guyenet PG, Kipnis J (2012) Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature 484:105–109. 10.1038/nature10907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubbelaar ML, Kracht L, Eggen BJL, Boddeke EWGM (2018) The kaleidoscope of microglial phenotypes. Front Immunol 9:1753. 10.3389/fimmu.2018.01753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzetti L, Menon R, Finardi A, Bergami A, Sica A, Martino G, Comi G, Verderio C, Farina C, Furlan R (2014) Activated macrophages release microvesicles containing polarized M1 or M2 mRNAs. J Leukoc Biol 95:817–825. 10.1189/jlb.0913485 [DOI] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma'ayan A, Chua WJ, Hansen TH, Turley SJ, Merad M, Randolph GJ (2012) Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 13:1118–1128. 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M (2010) Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330:841–845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, Rodewald HR (2015) Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518:547–551. 10.1038/nature13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, Glass CK (2014) Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159:1327–1340. 10.1016/j.cell.2014.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, Jaeger BN, O'Connor C, Fitzpatrick C, Pasillas MP, Pena M, Adair A, Gonda DD, Levy ML, Ransohoff RM, Gage FH, Glass CK (2017) An environment-dependent transcriptional network specifies human microglia identity. Science 356:eaal3222. 10.1126/science.aal3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M, Scott CL (2017) Does niche competition determine the origin of tissue-resident macrophages? Nat Rev Immunol 17:451–460. 10.1038/nri.2017.42 [DOI] [PubMed] [Google Scholar]
- Hagemeyer N, Hanft KM, Akriditou MA, Unger N, Park ES, Stanley ER, Staszewski O, Dimou L, Prinz M (2017) Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol 134:441–458. 10.1007/s00401-017-1747-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, Walker AJ, Gergits F, Segel M, Nemesh J, Marsh SE, Saunders A, Macosko E, Ginhoux F, Chen J, Franklin RJM, Piao X, McCarroll SA, Stevens B (2019) Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50:253–271.e6. 10.1016/j.immuni.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch UK, Kettenmann H (2007) Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci 10:1387–1394. 10.1038/nn1997 [DOI] [PubMed] [Google Scholar]
- Haynes SE, Hollopeter G, Yang G, Kurpius D, Dailey ME, Gan WB, Julius D (2006) The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci 9:1512–1519. 10.1038/nn1805 [DOI] [PubMed] [Google Scholar]
- Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, El Khoury J (2013) The microglial sensome revealed by direct RNA sequencing. Nat Neurosci 16:1896–1905. 10.1038/nn.3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G, Ginhoux F (2015) Ontogeny of tissue-resident macrophages. Front Immunol 6:486. 10.3389/fimmu.2015.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G, Ginhoux F (2018) Fetal monocytes and the origins of tissue-resident macrophages. Cell Immunol 330:5–15. 10.1016/j.cellimm.2018.01.001 [DOI] [PubMed] [Google Scholar]
- Jordão MJC, Sankowski R, Brendecke SM, Sagar, Locatelli G, Tai YH, Tay TL, Schramm E, Armbruster S, Hagemeyer N, Groß O, Mai D, Çiçek O, Falk T, Kerschensteiner M, Grün D, Prinz M (2019) Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363:eaat7554. 10.1126/science.aat7554 [DOI] [PubMed] [Google Scholar]
- Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, Wieghofer P, Heinrich A, Riemke P, Hölscher C, Müller DN, Luckow B, Brocker T, Debowski K, Fritz G, Opdenakker G, Diefenbach A, Biber K, Heikenwalder M, Geissmann F, et al. (2013) Microglia emerge from erythromyeloid precursors via pu.1- and Irf8-dependent pathways. Nat Neurosci 16:273–280. 10.1038/nn.3318 [DOI] [PubMed] [Google Scholar]
- Konishi H, Kobayashi M, Kunisawa T, Imai K, Sayo A, Malissen B, Crocker PR, Sato K, Kiyama H (2017) Siglec-H is a microglia-specific marker that discriminates microglia from CNS-associated macrophages and CNS-infiltrating monocytes. Glia 65:1927–1943. 10.1002/glia.23204 [DOI] [PubMed] [Google Scholar]
- Koso H, Tsuhako A, Lai CY, Baba Y, Otsu M, Ueno K, Nagasaki M, Suzuki Y, Watanabe S (2016) Conditional rod photoreceptor ablation reveals Sall1 as a microglial marker and regulator of microglial morphology in the retina. Glia 64:2005–2024. 10.1002/glia.23038 [DOI] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I (2014) Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159:1312–1326. 10.1016/j.cell.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ND, Hill JD, Juchem KW, Stefanopoulos DE, Modis LK (2014) RNA sequencing of microglia and monocyte-derived macrophages from mice with experimental autoimmune encephalomyelitis illustrates a changing phenotype with disease course. J Neuroimmunol 277:26–38. 10.1016/j.jneuroim.2014.09.014 [DOI] [PubMed] [Google Scholar]
- Li Q, Barres BA (2018) Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol 18:225–242. 10.1038/nri.2017.125 [DOI] [PubMed] [Google Scholar]
- Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M (2013) Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol 229:176–185. 10.1002/path.4133 [DOI] [PubMed] [Google Scholar]
- Mass E, Ballesteros I, Farlik M, Halbritter F, Günther P, Crozet L, Jacome-Galarza CE, Händler K, Klughammer J, Kobayashi Y, Gomez-Perdiguero E, Schultze JL, Beyer M, Bock C, Geissmann F (2016) Specification of tissue-resident macrophages during organogenesis. Science 353:aaf4238. 10.1126/science.aaf4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Sankowski R, Staszewski O, Böttcher C, Amann L, Sagar, Scheiwe C, Nessler S, Kunz P, van Loo G, Coenen VA, Reinacher PC, Michel A, Sure U, Gold R, Grün D, Priller J, Stadelmann C, Prinz M (2019) Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566:388–392. 10.1038/s41586-019-0924-x [DOI] [PubMed] [Google Scholar]
- Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada González F, Perrin P, Keren-Shaul H, Gury M, Lara-Astaiso D, Thaiss CA, Cohen M, Bahar Halpern K, Baruch K, Deczkowska A, Lorenzo-Vivas E, Itzkovitz S, et al. (2016) Microglia development follows a stepwise program to regulate brain homeostasis. Science 353:aad8670. 10.1126/science.aad8670 [DOI] [PubMed] [Google Scholar]
- Mrdjen D, Pavlovic A, Hartmann FJ, Schreiner B, Utz SG, Leung BP, Lelios I, Heppner FL, Kipnis J, Merkler D, Greter M, Becher B (2018) High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48:599. 10.1016/j.immuni.2018.02.014 [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, Ragozzino D, Gross CT (2011) Synaptic pruning by microglia is necessary for normal brain development. Science 333:1456–1458. 10.1126/science.1202529 [DOI] [PubMed] [Google Scholar]
- Prinz M, Priller J, Sisodia SS, Ransohoff RM (2011) Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat Neurosci 14:1227–1235. 10.1038/nn.2923 [DOI] [PubMed] [Google Scholar]
- Salter MW, Stevens B (2017) Microglia emerge as central players in brain disease. Nat Med 23:1018–1027. 10.1038/nm.4397 [DOI] [PubMed] [Google Scholar]
- Schaefer BC, Schaefer ML, Kappler JW, Marrack P, Kedl RM (2001) Observation of antigen-dependent CD8+ T-cell/ dendritic cell interactions in vivo. Cell Immunol 214:110–122. 10.1006/cimm.2001.1895 [DOI] [PubMed] [Google Scholar]
- Schafer DP, Stevens B (2015) Microglia function in central nervous system development and plasticity. Cold Spring Harb Perspect Biol 7:a020545. 10.1101/cshperspect.a020545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, Frampton J, Liu KJ, Geissmann F (2012) A lineage of myeloid cells independent of myb and hematopoietic stem cells. Science 336:86–90. 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- Scott CL, Zheng F, De Baetselier P, Martens L, Saeys Y, De Prijck S, Lippens S, Abels C, Schoonooghe S, Raes G, Devoogdt N, Lambrecht BN, Beschin A, Guilliams M (2016) Bone marrow-derived monocytes give rise to self-renewing and fully differentiated kupffer cells. Nat Commun 7:10321. 10.1038/ncomms10321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squarzoni P, Oller G, Hoeffel G, Pont-Lezica L, Rostaing P, Low D, Bessis A, Ginhoux F, Garel S (2014) Microglia modulate wiring of the embryonic forebrain. Cell Rep 8:1271–1279. 10.1016/j.celrep.2014.07.042 [DOI] [PubMed] [Google Scholar]
- Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M, Yamashita T (2013) Layer V cortical neurons require microglial support for survival during postnatal development. Nat Neurosci 16:543–551. 10.1038/nn.3358 [DOI] [PubMed] [Google Scholar]
- van de Laar L, Saelens W, De Prijck S, Martens L, Scott CL, Van Isterdael G, Hoffmann E, Beyaert R, Saeys Y, Lambrecht BN, Guilliams M (2016) Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity 44:755–768. 10.1016/j.immuni.2016.02.017 [DOI] [PubMed] [Google Scholar]
- Wlodarczyk A, Holtman IR, Krueger M, Yogev N, Bruttger J, Khorooshi R, Benmamar-Badel A, de Boer-Bergsma JJ, Martin NA, Karram K, Kramer I, Boddeke EW, Waisman A, Eggen BJ, Owens T (2017) A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J 36:3292–3308. 10.15252/embj.201696056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf SA, Boddeke HW, Kettenmann H (2017) Microglia in physiology and disease. Annu Rev Physiol 79:619–643. 10.1146/annurev-physiol-022516-034406 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrzavy T, Hametner S, Wimmer I, Butovsky O, Weiner HL, Lassmann H (2017) Loss of “homeostatic” microglia and patterns of their activation in active multiple sclerosis. Brain 140:1900–1913. 10.1093/brain/awx113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microglia E14-Specific Transcripts. List of 1321 transcripts exclusively expressed by Microglia at E14 (with no expression in Liver myeloid cells at E14 and a fold change equal to or higher than 5). Download Figure 2-1, XLSX file (112.2KB, xlsx)
Microglia P1-Specific Transcripts. List of 1168 transcripts exclusively expressed by Microglia at P1 (with no expression in Liver myeloid cells at P1 and a fold change equal to or higher than 5). Download Figure 2-2, XLSX file (99.1KB, xlsx)
E14 Liver Myeloid Cells-Specific Transcripts. List of 752 transcripts exclusively expressed by Liver myeloid cells at E14 (with no expression in Microglia at E14 and a fold change equal to or higher than 5). Download Figure 2-3, XLSX file (68.8KB, xlsx)
P1 Liver Myeloid Cells-Specific Transcripts. List of 855 transcripts exclusively expressed by Liver myeloid cells at P1 (with no expression in Microglia at P1 and a fold change equal to or higher than 5). Download Figure 2-4, XLSX file (76.7KB, xlsx)
Microglia Specific signatures during embryonic and early postnatal life. List of 759 Microglia Specific Transcripts both at E14 and P1. Download Figure 2-6, XLS file (130.5KB, xls)
List of 94 Microglia conserved transcripts not present in liver myeloid cells but always expressed by microglia at E14, P1 and in adulthood. Download Figure 3-1, XLS file (35.5KB, xls)
65 Microglia Specific Transcripts Throughout Life. List of 65 homeostatic microglia-specific transcripts, which are always expressed by microglia throughout life at E14, P1and in adulthood, but neither expressed in liver macrophages at E14 and P1 nor in adult peritoneal macrophages at steady state or after differentiation towards an M1 or M2 phenotype. Download Figure 3-2, XLSX file (22KB, xlsx)