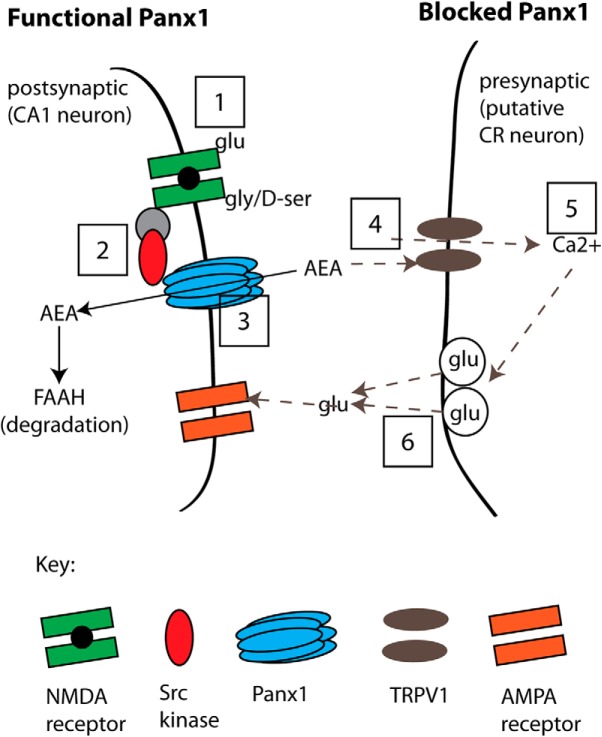
Figure 11.
A model of the proposed pathway for suppression of glutamate release by postsynaptic metabotropic NMDAR signaling to Panx1. Left, “Functional Panx1,” the scenario when Panx1 channels are operating normally. This physiological state is indicated as Steps 1–3 (boxed numbers) where the binding of glutamate and glycine/d-serine to NMDARs (Step 1) initiates Src-mediated phosphorylation of Panx1 (Step 2), which facilitates clearance of AEA (Step 3) for degradation by FAAH. Our evidence for this sequence is that blocking ligand binding to NMDRs (with d-APV+CGP; at Step 1) but not postsynaptic increases in Ca2+ (with BAPTA), or blocking Src activation of Panx1 (with TAT-Panx308 or PP2; at Step 2), or inhibiting Panx1 (with α-panx1, 10panx, or Panx1−/−; at Step 3) all augmented sEPSC frequency. Right, “Blocked Panx1,” the sequence of events occurring when Panx1 or metabotropic NMDARs have been blocked (Steps 4–6; dashed arrows). In the current study, direct block of Panx1 increases tissue AEA levels (Step 4), and we propose that AEA acts on presynaptic TRPV1 to increase Ca2+ (Step 5) and facilitate activity-dependent glutamate release (Step 6). Our evidence for this sequence is that facilitated glutamate release was prevented by TRPV1 block (with CPZ or in TRPV1−/−; at Step 4), or by bath-applied EGTA-AM (at Step 5). We also quantified TRPV1 expression in the hippocampus (PCR, qPCR) and found it at presynaptic sites (by immuno-EM) and propose that these are CR cells synapsing onto CA1 neurons because CR cells are the only confirmed site of TRPV1 expression.