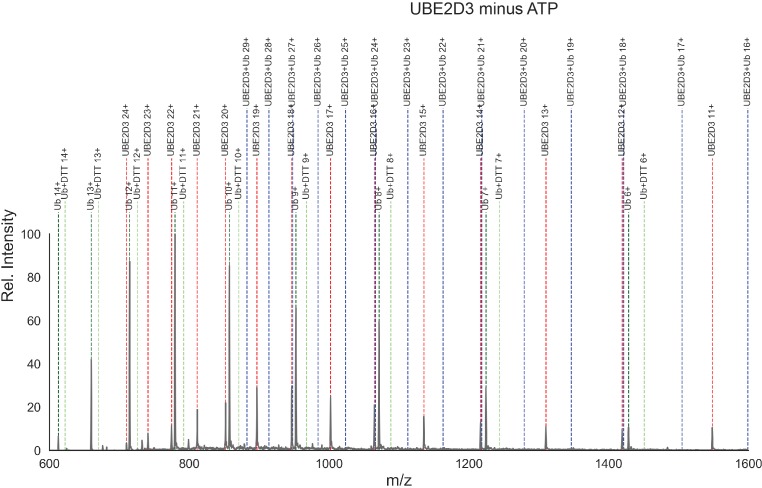
Appendix 2—figure 2. Mass spectra for control ubiquitin loading reaction containing UBE2D3.
Mass spectrum for a negative control ubiquitin charging reaction containing UBE2D3 (10 µM), E1 (1 µM) and wild-type human ubiquitin (15 µm) in reaction buffer without ATP (30 mM Tris, pH 7.5, 100 mM NaCl, 5 mM MgCl2, and 1 mM DTT). The reaction components were incubated at room temperature for 5 min, then quenched by adding 90 μL of 5% acetic acid. The detection of proteins was performed on an Agilent LC-MSD. Mass spectra were acquired in positive-ion mode, scanning from 500 to 1700 m/z. The electrospray voltage was set to 4 kV and the gas temperature in the spray chamber was maintained at 350°C. A stationary phase, Zorbax 300SB C3 150 mm ×2.1 mm column was used for separation (Agilent). The mobile phase A buffer was 0.2% formic acid, and the mobile phase B was 0.2% formic acid with 10% methanol and 90% acetonitrile. The flow rate was 0.2 ml min−1. After a 25 min delay, the effluent was directed into the mass spectrometer. Linear gradients started with 5% mobile phase B and finished at 95% from 25 to 50 min. Data were processed using the ChemStation software package. Deconvolution of the spectra into the observed species and their abundances is shown in Appendix 2—figure 2—source data 1 . Dotted blue lines are shown where peaks would be expected for UBE2D3 ~ ubiquitin. Notice that none were found in the absence of ATP.