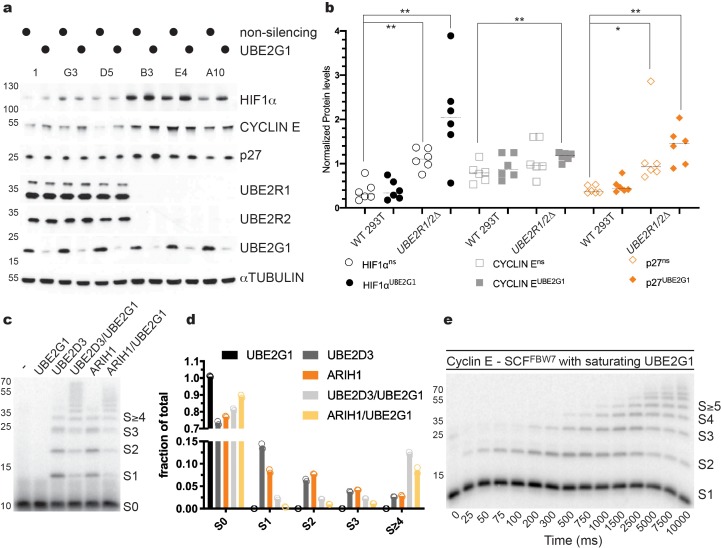
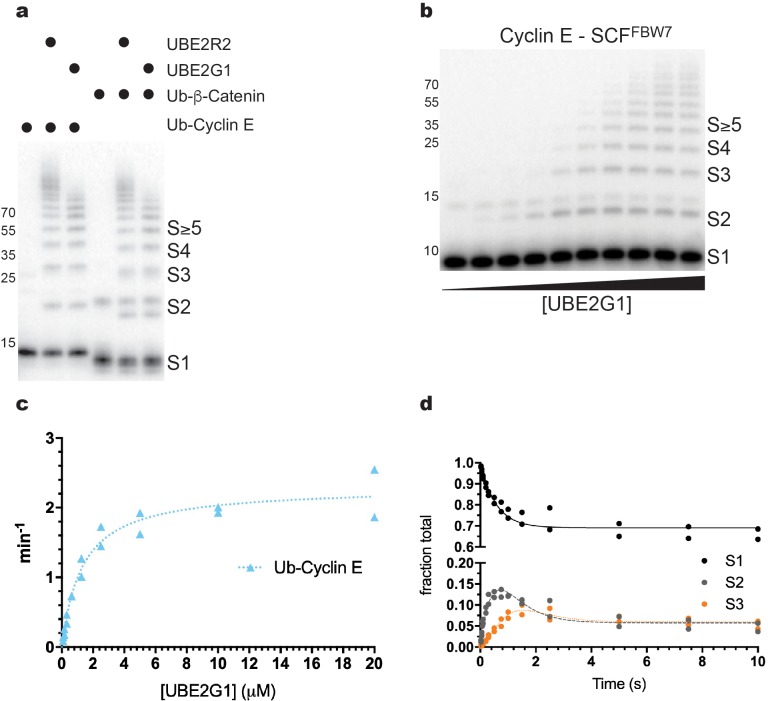
Figure 7. UBE2G1 functions redundantly with UBE2R1/2 in cells and in SCF enzyme reactions in vitro.
(a) Comparison of the steady state stabilities of HIF1α, CYCLIN E, and p27 proteins in HEK 293T control or UBE2R1/2 double knockout cells treated with siRNA targeting UBE2G1 expression. (b) Quantitative comparison of the steady state levels for HIF1α (open or closed black circles), CYCLIN E (open or closed gray squares), and p27 protein (open or closed orange diamonds). P-values were calculated using an unpaired t-test (* and ** denote values of less than 0.05 or 0.01, respectively). P-values for all relevant combinations are provided in Figure 7—source data 1. Three biological replicates each for control and UBE2R1/2 double knockout cells were used to generate the figure with duplicate technical replicates. (c) Representative autoradiogram of a Cyclin E peptide ubiquitylation reaction with either ARIH1, UBE2D3, or UBE2G1 alone or in combination. (d) Graphical representation of the levels of unmodified substrate and ubiquitylated products from the reactions shown in panel c. Duplicate technical replicates were performed to generate the figure. Source data have been provided in Figure 7—source data 2 for panel d. (e) Representative autoradiogram of a Cyclin E ubiquitylation reaction with UBE2G1 levels (12.5 μM) sufficient to saturate the SCF complex. Figure 7—figure supplement 1d shows the fit of the data to the kinetic model. The enzyme concentrations have been provided in Supplementary file 2.