Fig. 4. Distinct gene expression profiles of SCCs reveal prognostic genes.
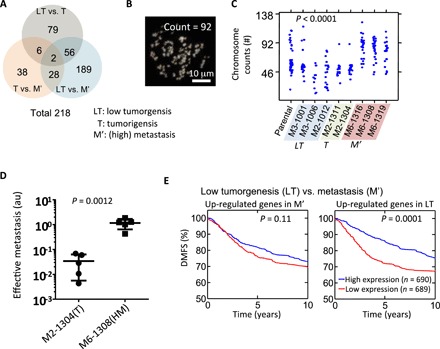
(A) Venn diagram showing the number of genes that are found to be significantly different (>5-fold and P value from one-way ANOVA <0.05) between three different in vivo grades of aggressiveness for SCCs (i.e., LT versus T, T versus M′, and LT versus M′). M′ includes both M and HM. (B and C) Representative image showing 4′,6-diamidino-2-phenylindole (DAPI)–stained spreading chromosome of SCC-M6-1308 (B). Chromosome number counted using the metaphase spreading assay for parental cells (n = 44), and cells from SCC-M3-1001 (n = 24), SCC-M3-1006 (n = 11), SCC-M2-1012 (n = 22), SCC-M2-1311 (n = 18), SCC-M2-1304 (n = 18), SCC-M6-1316 (n = 26), SCC-M6-1308 (n = 31), and SCC-M6-1319 (n = 22). One-way ANOVA test shows there is a significant difference, with a P < 0.0001 (C). (D) Score for effective metastasis to the lung in the tail-vein injection mouse model (n = 5) shows significant difference (P = 0.0012 by Student t test) between tumorigenic clone SCC-M2-1304 (mean lung effective metastasis score, 0.034) and metastatic clone SCC-M6-1308 (1.159). (E) Differentially expressed genes between LT SCC versus M′ SCC were used to investigate their prognostic power. A cohort of 1379 tumors from patients with breast cancer was used to test the predictive potential of identified gene sets. Patients were separated into two groups based on the average expression level of these identified genes, and the Kaplan-Meier survival curves for the two groups of patients were plotted. For the genes that were up-regulated in the M′ SCCs, no significant prognostic effect was found. However, the results show that patients with higher expression levels of metastasis suppressor genes (i.e., up-regulated genes in LT) have a significantly longer survival time than those with low expression (P = 0.0001). P value is evaluated using log-rank test.
