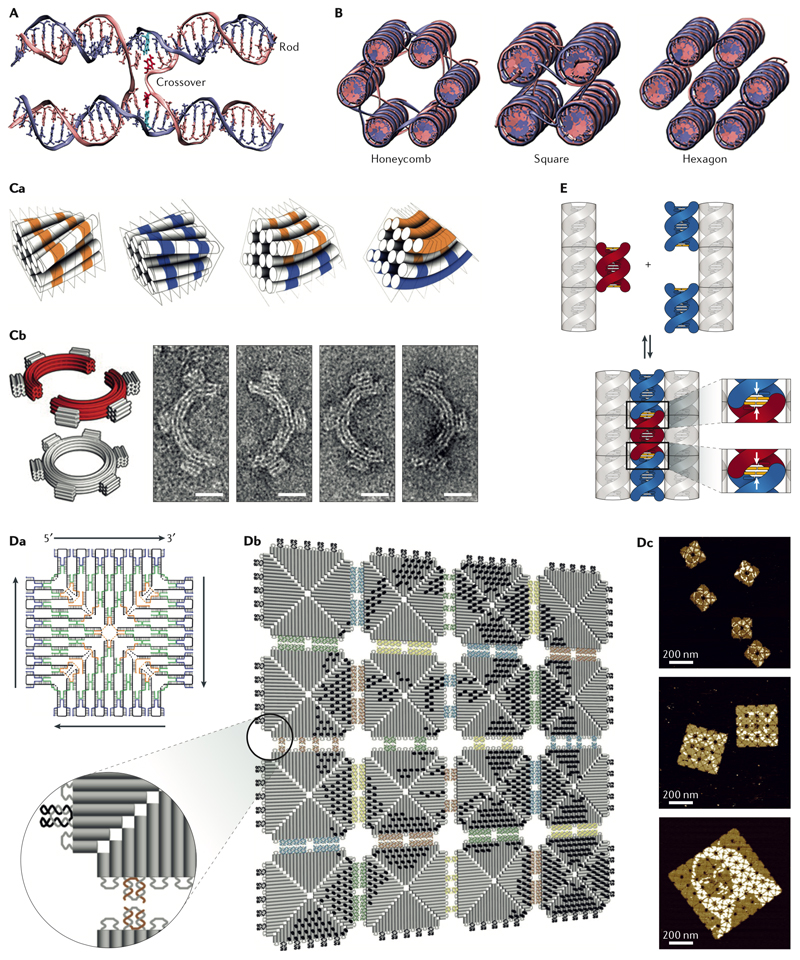
Fig. 1. Design principles of DNA origami structures and higher-order self-assembly using origami tiles.
A| The schematic of an antiparallel Holliday junction forming a double crossover between two adjacent helices in a honeycomb DNA origami six-helix bundle. Note, how the nucleobases at the crossover position (highlighted in red and cyan in the left crossover) still maintain their natural angle for duplex formation. B| The honeycomb (left), square (middle), and hexagonal (right) helical packings are the most common helical arrangements. The crossovers were deleted in the hexagonal packing due to multiple possible segment lengths and scaffold routings. C| Curvature design through base pair deletion or insertion. Ca| The base deletion (in orange) with a segment size smaller than 7 bp in the honeycomb arrangement causes a left-handed twist whereas a base insertion (in blue) results in a right-handed twist. Calculated combination of these two types of twists can be used to create curvature without causing marked global deformation. Ca| Negative-stain transmission electron microscopy (TEM) micrographs of two curved half-circles origami monomers and their six-tooth gear dimer (scale bar 20 nm)43. D| Fractal assembly. Da| The strand map of the origami tile used for fractal assembly. The staples are shown in blue, green, and yellow and the scaffold in black. Db| Fractal assembly of DNA origami tiles using edge loops and 2-nucleotide staple hybridization. The Mona Lisa’s pattern could be printed by adding double-stranded extensions to the selected staples on the surfaces of the tiles55. Dc| Atomic force microscopy confirmed the correct assembly of the tiles. E| Click contacts. Association of shape-complementary surface features can be driven by base stacking57.
Panel D adapted from REF. 55, with permission from Springer Nature.