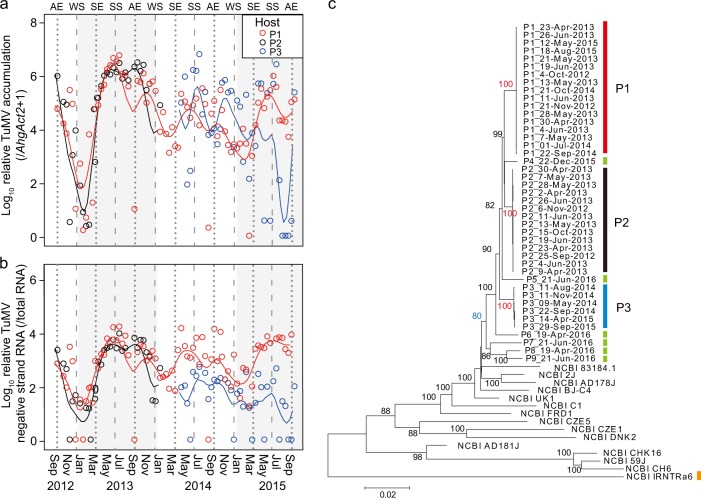
Fig. 2.
Three-year observation on within-host accumulation and phylogenetic relationship of TuMV in three infected Arabidopsis halleri plant patches. Temporal changes of accumulation (a) and negative-strand RNA (b) of TuMV in upper leaves of plant patches 1, 2, and 3 (P1, P2, and P3, respectively). Solid lines indicate smooth-spline fittings. Negative-strand RNA was detected by strand-specific RT-qPCR. Broken lines and dashed lines indicate spring/autumn equinoxes (SE/AE) and summer/winter solstices (SS/WS), respectively. The year periods (i.e., 2012, 2013, 2014, and 2015) were shown by white and grey shadings. c Phylogenetic relationships between TuMV strains detected from the three patches (red, black, and blue bars represent P1, P2, and P3, respectively) during the 3-year observation. One-time sample from additional six plants were included (P4‒P9; indicated by green coloured bars). Sample IDs were coded by plant patches and sampling dates. Previously reported five, four, and four strains selected from Japan, continental Asia, and Europe, respectively, were included, and the sequence of TuMV basal-B strain (IRNTRa6; whose original host is Rapistrum rugosum) was used as an outgroup (indicated by an orange coloured bar). The bootstrap values (% of 1000 replicates) are listed next to the branches. The bootstrap values in red supported the monophyly of virus sequences within plant patches 1, 2, and 3, and the value in blue supported the monophyly of virus sequences from the studied population