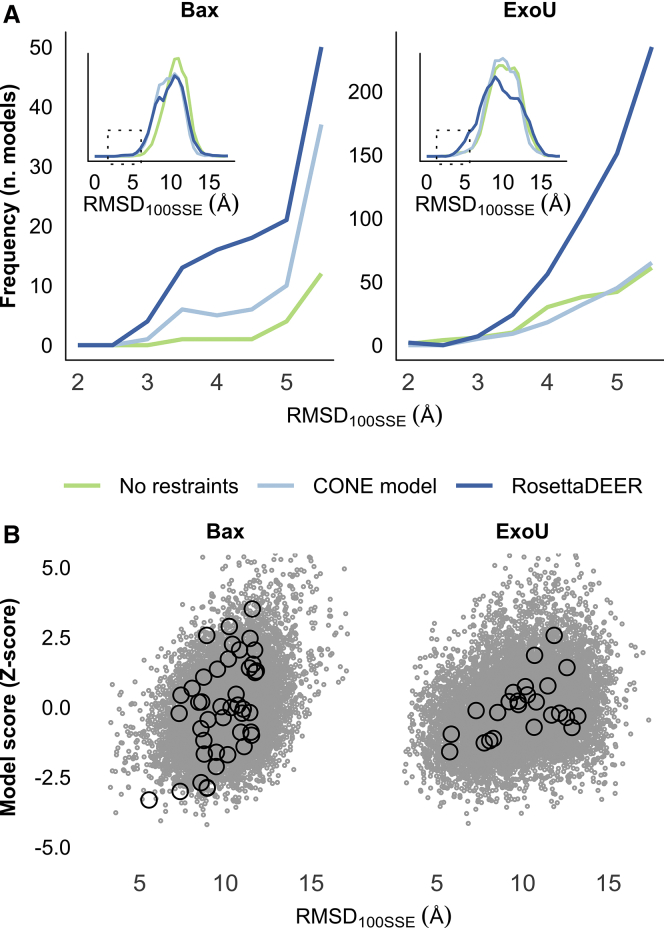
Figure 4.
Structure prediction of Bax and ExoU using DEER decay data. (A) De novo protein folding of native-like models using DEER decay restraints with RosettaDEER, Cβ-Cβ distance restraints with the CONE model, or no restraints. Inset: spread of all models generated using these three methods. (B) Accuracy of de novo folded models (gray dots) and clusters (black circles) as a function of combined DEER and Rosetta Z-score.