Figure 1.
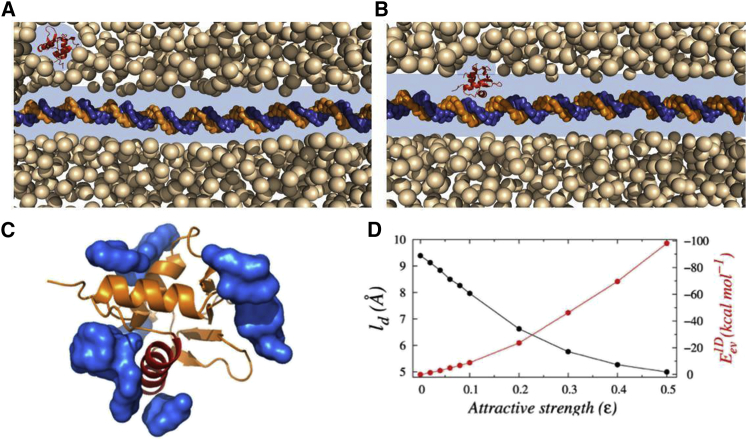
A schematic representation of the target search process of DNA-binding proteins (DBPs) inside a crowded medium. (A) The crowding agents form depletion zones (highlighted as the blue region) around the protein and DNA molecules separately when they are far from each other (3D diffusion). (B) When the molecules are in close proximity, the depletion regions merge, and the protein moves along the DNA contour one dimensionally through the composite depletion zone. It should be noted that the width of the depletion region is not to scale. In the absence of a searching protein on the DNA surface, the nearest crowder to the DNA locates approximately its radius distance away. The average of such distances provides the estimation of width of the depletion region, shown in (D). The crowder molecules in the front and back of the DNA are removed intentionally to show the depletion region clearly. (C) During both 3D and 1D diffusion, the crowder molecules interact with the diffuser protein through nonspecific short-range attractive forces, as can be seen from the preferential association of PEG600 crowders (shown in the blue surface) on Sap-1 protein surface in an all-atom simulation. In this context, it is worth mentioning that PEG molecules are often considered as inert crowding agents and their influence is described by volume exclusion only. However, results in (C) suggest the presence of a nonspecific attractive interaction between the protein and PEG molecules, which should be taken into consideration while explaining its action. (D) Variations in width of the depletion region (width, ld) around the DNA molecule and excluded volume interactions between the searching protein and the crowder molecules with the increasing attractive strength of crowders (ε) are shown. To see this figure in color, go online.