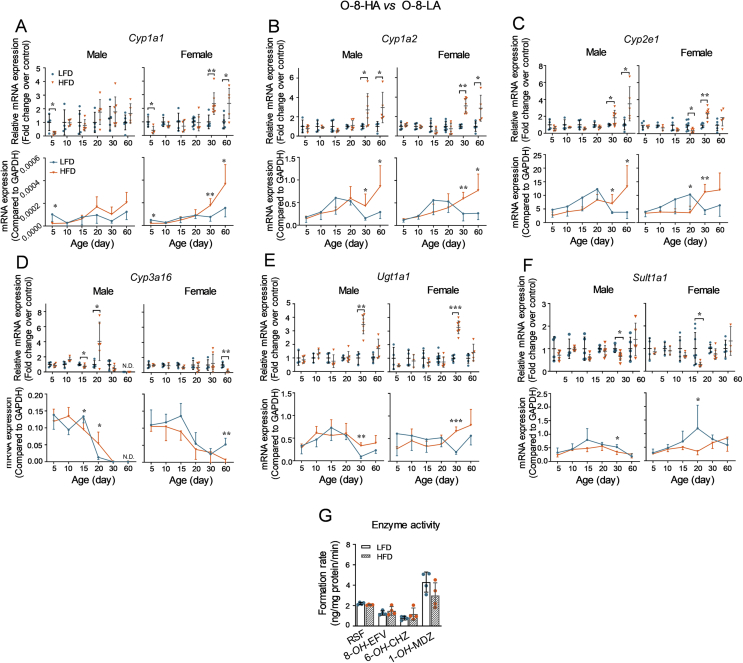
Figure 5.
Effects of parental HFD on the ontogenic expression patterns of DMEs in offspring liver. The offspring derived from parental mice fed with a LFD or HFD for 8 weeks were sacrificed at different ages after birth (n = 3–6 per group). The liver tissues were collected to determine the mRNA expression levels and enzyme activities of DMEs. (A)–(F) The mRNA expression levels were measured using the RT-qPCR method and normalized to Gapdh. The upper panels (relative mRNA expression levels) were calculated using the 2−ΔΔCt method compared to the LFD groups. The lower panels (mRNA expression levels) were expressed as 2–[Ct(DMEs) − Ct(Gadph)]. (G) Enzyme activities of CYP1A, 2B, 2E, and 3A in four random selected male offspring at day 30 from the LFD or HFD group were determined by the UPLC–QTOFMS method and expressed as the formation rates of RSF, 8-OH-EFV, 6-OH-CHZ, and 1-OH-MDZ, respectively. Data are depicted as mean ± SD. Each dot represents a value from one mouse. Statistical significance between two groups was determined by an unpaired Student's t-test. *P < 0.05, **P < 0.01, ***P < 0.001, HFD versus the age-matched LFD groups.