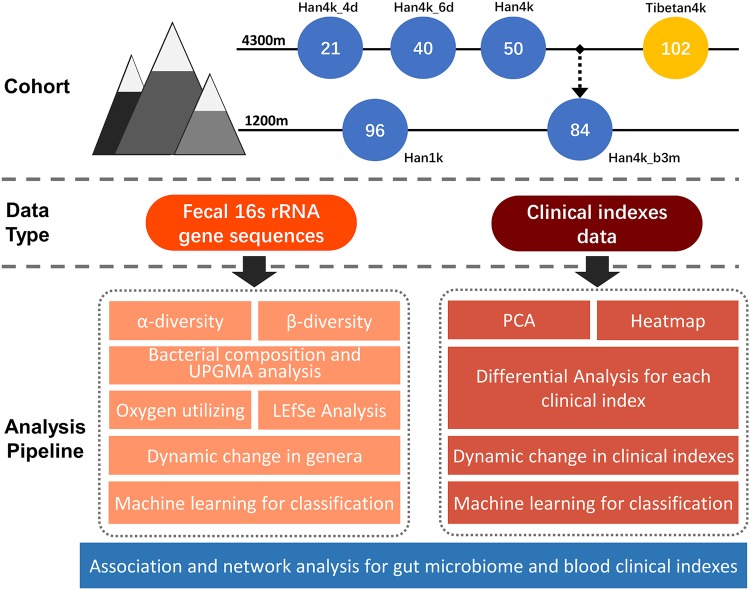
FIG 1.
Overview of the analysis pipeline. The fecal and blood samples were collected and analyzed. The fecal samples were subjected to 16S rRNA gene sequencing followed by diversity analysis, bacterial composition analysis, functional prediction, and differential analysis. The blood samples were analyzed with 76 clinical indexes. The clustering analysis and statistical tests were applied to the clinical indexes. An XGBoost-based machine learning model was used to distinguish the different groups based on the bacterial and clinical indexes. Finally, analysis of the association and network between the gut microbiota and clinical indexes were used to illustrate the correlation and group-specific signatures.