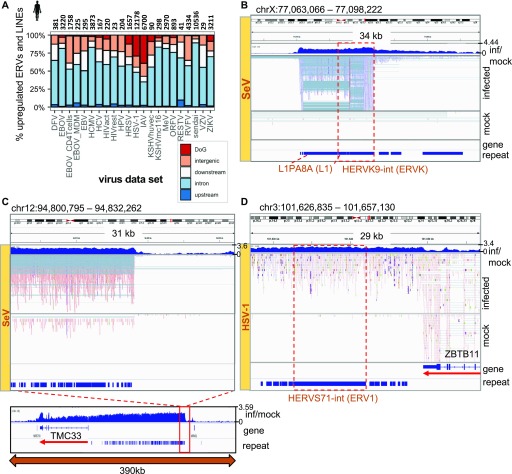
Figure 3. Relationship between differentially up-regulated TEs and other transcriptional events.
(A) Significantly up-regulated TEs and their relationship with genes across human virus data sets. Total numbers are indicated above each bar. Upstream and downstream regions are defined as 3 kb from the transcriptional start site or termination site, respectively. Intergenic refers to an element that is greater than 3 kb from any annotated ENSEMBL gene boundaries. (B, C, D) Genome browser view showing (B) intergenic TEs that are highly up-regulated in SeV-infected cells, (C) up-regulation of a stretch of hundreds of TE and repeat elements ∼100 kb upstream of TMC33 gene during SeV infection, and (D) TR of the ZBTB11 gene into HERVS71-int (ERV1) in HSV-1-infected cells. “inf/mock” track shows the log2(RPKM-normalized coverage of infected over mock samples). “repeat” and “gene” tracks show DFAM repeat and RefSeq gene model annotations, respectively.