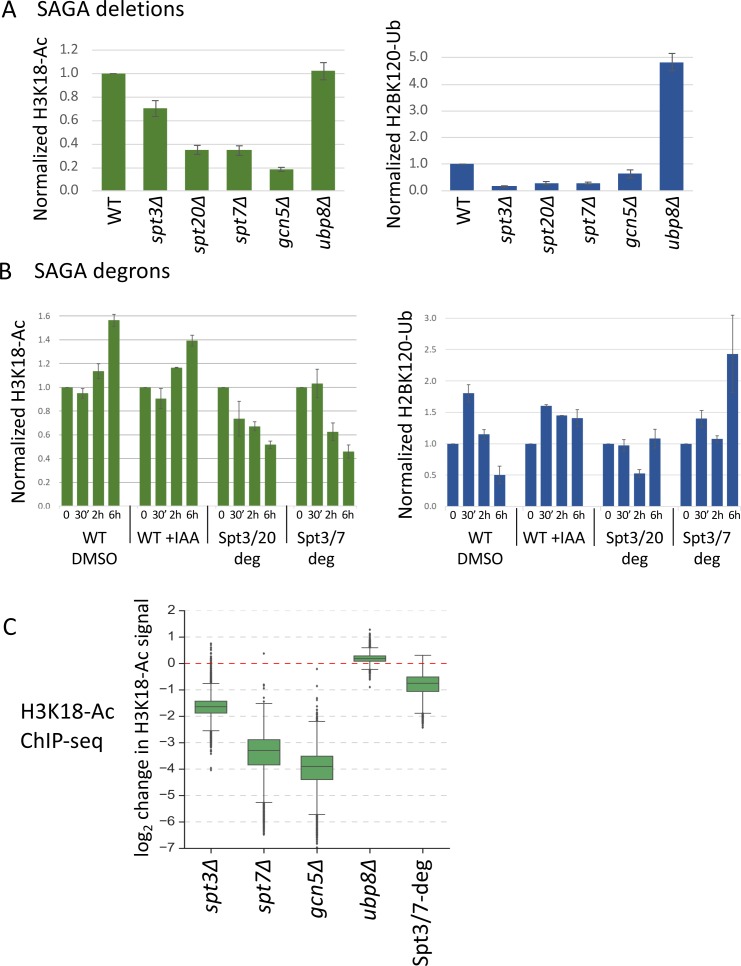
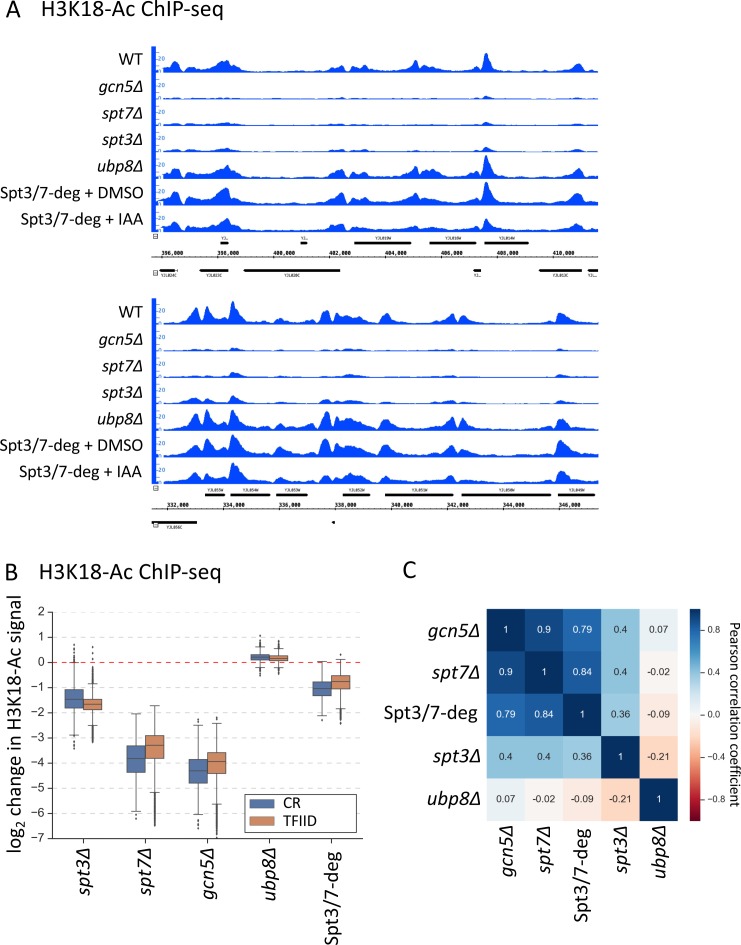
Figure 5. Histone modifications in SPT deletion and degron strains.
(A, B) Quantitation of total H3K18-Ac and H2B-Ub in the SPT deletion and degron strains, normalized to the TFIIH subunit Tfg2 as a loading control. Data are from Figure 5—figure supplement 1. In panel (B), time after IAA or DMSO addition is indicated. (C) Box plot showing the ChIP-seq assayed log2 change per gene in H3K18-Ac signal in the SAGA deletion mutants and Spt3/7 degron strain. Degron strain was treated for 30 min with IAA before formaldehyde crosslinking. Mean values from replicate experiments are plotted.