Abstract
While the ‘Micro-Tom’ TILLING mutant library is used for a wide range of purposes, including both basic research of gene function and breeding of commercial cultivars, genome-wide distribution and frequency of mutations have not yet been thoroughly elucidated on a population scale. In this study, we developed a 96-plex exome sequencing method to identify and analyze mutations within the TILLING mutants that were developed in the University of Tsukuba. First, an Illumina paired-end sequencing coupled with 96-plex exome capture resulted in the acquisition of an exome sequence dataset with an average read count of 5.6 million for the 95 mutants. Over 98% of the capture target region could be covered by the short reads with an averaged read depth of 12.8, which enabled us to identify single nucleotide polymorphisms and Indels in a genome-wide manner. By subtracting intra-cultivar DNA variations that are present between wild-type ‘Micro-Tom’ lines, we identified 241,391 mutation candidates in 95 mutant individuals. Of these, 64,319 and 6,480 mutations were expected to cause protein amino acid substitutions or premature stop codon, respectively. Based on the exome mutation dataset, a mutant line designated ‘TOMJPW601’ was found to carry a premature stop codon mutation (W261*) in a putative auxin influx carrier gene SlLAX1 (Solyc09G014380), consistent with our previous report of its curly leaf phenotype. Our results suggested that a population-scale mutation database developed by multiplexed exome sequencing could be used for in silico mutant screening, which in turn could contribute to both gene function research and breeding programs.
Keywords: 96-plex exome sequencing, in silico mutant screening, Micro-Tom TILLING library, mutation database, tomato
Introduction
Tomato (Solanum lycopersicum L.) is one of the most economically important horticultural crops in the world; it is a source of vitamins, minerals, and other substances promoting health. While new breeding technologies such as CRISPR/Cas-mediated genome editing are rapidly emerging as a tool for efficient crop breeding, random mutagenesis technologies such as TILLING are still important in both basic scientific research and crop breeding (Ito et al. 2015; Okabe et al. 2011; Shimatani et al. 2017). In tomato, a TILLING mutant library has been generated using some cultivars such as ‘M82’, ‘Red Setter’, and ‘TPAADASU’ by chemical mutagens (e.g., ethyl methanesulfonate) or gamma-ray irradiation (Gady et al. 2009; Minoia et al. 2010; Piron et al. 2010). In particular, a dwarf tomato cv. ‘Micro-Tom’ has become available as a tomato model cultivar for the development of a functional genomic tool with useful traits and benefits for research (small plant size, short life cycle, useful databases for examining the genome information and gene function) (Kobayashi et al. 2014; Meissner et al. 1997; Okabe et al. 2011, 2013; Shirasawa et al. 2016; Sun et al. 2006; Watanabe et al. 2007). For the development of such a functional tool, we have generated a mutant population based on ‘Micro-Tom’ by using EMS or gamma-ray irradiation under National BioResource Project (NBRP)-Tomato as previously reported (Matsukura et al. 2007; Saito et al. 2011; Shikata et al. 2015; Watanabe et al. 2007). These induced mutants were used for both forward and reverse genetic studies. In forward genetics, they have been used to identify genes responsible for important agricultural traits such as stress tolerance and parthenocarpy (Gauffier et al. 2016; Mazzucato et al. 2015). By contrast, in reverse genetic screening, they were used to show the function of specific genes important for the accumulation of health-promoting substances like gamma-amino butyric acid (GABA) or vitamin C (Baldet et al. 2013; Takayama et al. 2017).
There are several methods to screen for a specific mutant in a gene of interest. A gel electrophoresis-based method using CEL-1 endonuclease is the most used traditional screening method whereby mutations are manually screened according to the difference in gel electrophoresis (Eliot et al. 2008; Greene et al. 2003). High-resolution melting (HRM) is a post-PCR screening method that uses a quantitative PCR instrument, whereby mutations are screened based on the difference in the melt curve between homogeneously and heterogeneously hybridized DNA molecules. HRM has been demonstrated to be an efficient mutant screening method in several crops (Dong et al. 2009; Lochlainn et al. 2011; Takagi et al. 2018; Yano et al. 2017, 2018). In both the CEL-1 based method and HRM, changes in DNA bases are required to be confirmed by DNA sequencing (e.g., sanger method) after the initial screening. Recently, an NGS-based amplicon sequencing method has been also used to identify mutations in TILLING populations (Gupta et al. 2017; Rigola et al. 2009; Tsuda et al. 2015; Zhu et al. 2012). Several time-consuming steps (e.g., PCR amplification of target DNA fragments and subsequent multiplexing) are required in a traditional target amplicon sequencing method; however, the recently developed single Primer Extension Technology (SPET) method has enabled efficient construction of a highly-multiplexed NGS library (e.g., 384-plex) (Scaglione et al. 2019).
Theoretically, whole genome NGS can be a robust mutation identification method as it enables large scale identification of genome-wide mutations. Once such genome-wide mutation information is obtained, computational search and identification of a specific mutation are possible. However, even though the cost of whole genome NGS has been reducing, it is still expensive, especially when analyzing mutants on a population scale. In contrast to whole genome NGS, exome sequencing obtains and analyzes DNA sequences of only exonic (protein-coding) regions (Baker 2012; Neves et al. 2013; Valdés-Mas et al. 2012). In exome sequencing, exonic genomic regions are captured by hybridization using chemically modified DNA probes prior to sequencing. In tomato, the total size of the exonic genomic region is expected to be around 50 to 60 Mb according to the Heinz1706 genome reference, while the size of the whole genome is expected to be more than 900 Mb (Aoki et al. 2010; Kobayashi et al. 2014; The Tomato Genome Consortium 2012; Sol Genomics Network, https://solgenomics.net). Thus, target sequence length becomes much smaller in exome sequencing than whole genome NGS, which reduces experimental costs.
In this study, we developed a 96-plex exome capture method to enable exome sequencing at population scale in the ‘Micro-Tom’ TILLING mutant population. While multiplexing is possible for up to 24 in a commercially available exome capture experimental kit (e.g., Roche SeqCapEZ®), we extended the SeqCapEZ method to enable 96-plex exome sequencing. By using several lines of wild-type control NGS data to subtract intra-cultivar DNA variations, we could identify a total of 241,391 mutation candidates in 22,353 genes in 95 independent mutants. Our results also suggest that a large population-scale mutation database might enable us to conduct in silico screening of specific mutants carrying mutation(s) in a gene of interest.
Materials and methods
Plant materials and growth conditions
TILLING mutant population had been previously been developed using chemical mutagen EMS and gamma-ray irradiation as reported previously (Saito et al. 2011; Shikata et al. 2015). Seeds of wild-type tomato cultivar ‘Micro-Tom’ that were used to develop the TILLING mutant library were derived from the Kazusa DNA Research Institute in Japan. This wild-type strain is registered as ‘TOMJPF00001’ in the NBRP Tomato database (TOMATOMA; http://tomatoma.nbrp.jp/). The ‘TOMJPF00001’ strain had been subjected to six generations of self-fertilization based on the original ‘Micro-Tom’ line of Scott and Harbaugh 1989. All of wild-type ‘Micro-Tom’ lines used in this study were labelled or provided as the ‘TOMJPF00001’ strain. However, there was a possibility that these wild-type ‘Micro-Tom’ had been genetically segregated from the original ‘TOMJPF00001’ strain and independently fixed through self-fertilization over the years by independent persons.
There are three kinds of mutant types available in University of Tsukuba; ‘TOMJPW’, ‘TOMJPE’ and ‘TOMJPG.’ They had been subjected to two rounds of EMS treatment (TOMJPW), a one-time EMS treatment (TOMJPE), or gamma-ray irradiation (TOMJPG), respectively (Saito et al. 2011; Shikata et al. 2015). For isolation of genomic DNA and construction of multiplexed exome library, plants of wild-type ‘Micro-Tom’ and 95 mutant lines were grown under greenhouse conditions at the University of Tsukuba. These mutants included 68 ‘TOMJPW’ lines, 22 ‘TOMJPE’ lines, and five ‘TOMJPG’ lines (Supplementary Table S1). Prior to this study, seeds of these mutant lines had been already obtained. Briefly, seeds of M3 generation (TOMJPE and TOMJPG lines) or M3M3 generation (TOMPW lines) had been obtained by self-fertilization of one M2 (TOMJPE and TOMJPG lines) or M3M2 (TOMPW lines) plant that showed some morphological changes relative to the wild type. Then, if necessary, seeds of M4–7 generation (TOMJPE and TOMJPG lines) or M3M4 generation (TOMPW lines) were maintained as bulked seeds by combining self-fertilized seeds of 10 parental plants. Thus, mutant lines used in this study were not isogenic lines. In each mutant, one to ten siblings were grown to obtain a bulked leaf sample. Either of M4–7 generation (TOMJPE and TOMJPG lines) or M3M3–4 generation (TOMJPW lines) was used in this study because seeds of earlier generations were no longer available in some mutant lines. In some mutant lines, it was also unable to prepare 10 plants because it was difficult to germinate seeds or grow plants probably due to the large number of induced mutations in their genomes. Bulked samples were frozen in liquid nitrogen and stored at −80°C until use.
Isolation of genomic DNA and genome-tip® column purification
Frozen leaf samples were ground to powder with the QIAGEN TissueLyser® (QIAGEN, Venlo, Netherlands), and genomic DNA was isolated with the Maxwell® 16 tissue DNA isolation kit according to the manufacturer’s protocol. Then, DNA samples were purified with QIAGEN Genome-tip® 20/G columns to eliminate contaminations (e.g., polysaccharide and RNA). The concentrations of DNA were determined using Nanodrop® (Thermo Scientific, MA, USA), and the DNA samples were used for 96-plex exome library preparation or whole genome Illumina sequencing.
Library preparation for 96-plex exome sequencing
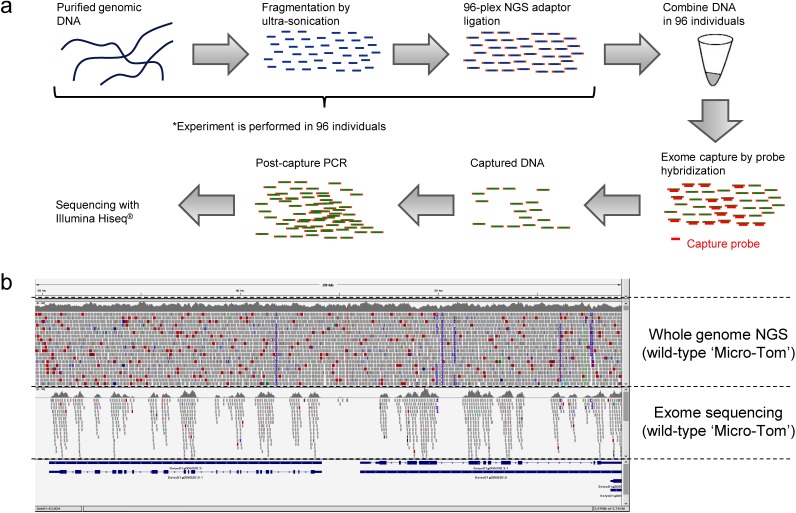
To construct 96-plex exome sequencing library, we took an approach that combines NEBNext® Ultra DNA Library Prep Kit for Illumina (New England Biolabs Inc., Ipswich, MA, USA), NEBNext® Multiplex Oligos for Illumina (Dual Index Primers Set 1) (New England Biolabs Inc.), and Roche-NimbleGen SeqCapEz® exome library preparation kit (Roche, Basel, Switzerland) (summarized in Figure 1). First, genomic DNA was subjected to ultra-sonication with Covaris S220® (Covaris, Inc., MA, USA), sheared DNA was then reacted with the ‘End repair and preparation solution’ followed by ligation with Illumina-compatible adaptor provided by NEBNext® library preparation kit. After DNA fragment size selection with AMPure® XP Beads (Beckman Coulter, Inc., Brea, CA, USA), adaptor-mediated PCR was performed to attach 96-plex index sequences with the following reactions; 98°C for 30 s for initial denature, six cycles of 98°C for 10 s and 65°C for 75 s for DNA amplification, 65°C for 5 min for extension, 4°C for holding. The lengths of amplified DNA fragments were 250–500 bp including adaptor sequences according to the analysis with Agilent 2100 DNA analyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). After further purification with AMPure® XP Beads, equal amounts of DNA were taken from each sample and then combined in a new tube.
Figure 1. Exome sequencing in the ‘Micro-Tom’ mutant population. (a) A simplified cartoon illustrating a procedure of 96-plex exome capture. (b) A bowtie2 alignment examples of whole genome NGS reads (upper side) and exome sequencing (lower side). Both data were derived from wild-type ‘Micro-Tom.’.
Next, combined DNA was subjected to exome capture treatment using the customized tomato exome probe set that was developed in a previous study (Pulungan et al. 2018). This exome probe set was designed based on the tomato genome reference Heinz1706 SL2.5 (Sol Genomics Network, https://solgenomics.net) to capture the exonic regions of 34,768 tomato genes (total target length=49.5 Mb). To enable 96-plex exome capture, we designed and used customized hybridization enhancing (HE) oligos instead of those provided by SeqCapEz® kit (Supplementary Table S2). These HE oligos were required to suppress capturing of non-target sequences. Except for using the customized HE oligos, all steps of exome capture experiment were performed according to the version 5.0 protocol of the Roche SeqCapEz® kit. To evaluate the capture efficiency of target DNA, we performed quantitative PCR in both pre-capture and post-capture DNA samples with Thunderbird® SYBR qPCR Mix (Toyobo, Osaka, Japan) using the primers of three control genes: Solyc06g082600 (5′-CAG ATC CAA ACC CAG ACG AC-3′ and 5′-TCC AGC TAC GAG CAG TGG TC-3′), Solyc10g080500 (5′-GGC TGT CTT CCC TAG TAT TGT G-3′ and 5′-GTT AAG AGG AGC TTC TGT GAG G-3′), and Solyc08g062800 (5′-GAT ATT TTC CAA TTG CTG CCA CCA AAG-3′ and 5′-CTC ATC ACG CTT CAC AAG GAT C-3′). The abundance of DNA fragments was quantified based on the standard calibration curve that was obtained using different concentrations of genomic DNA, then enrichment efficiency of exome capture was calculated based on DNA fragment abundance. We confirmed that enrichment efficiency was more than 10-fold in all of the three control genes. The resultant post-capture DNA was further subjected to Illumina Hiseq-2000 sequencing with dual index 100 bp paired-end sequencing mode using the outsourced service of Macrogen Japan Corp. (Kyoto, Japan). Demultiplexed sequence datasets were obtained as fastq files and can be found in a public database (DDBJ accession No. DRR184778-DRR184873).
Whole genome resequencing in wild-type ‘Micro-Tom’
As described above, it was suspected that there were several independent lines for the wild-type ‘Micro-Tom’ although they were all labeled or provided as the ‘TOMJPF00001.’ Thus, whole genome sequencing data was newly obtained in three independent lines of wild-type ‘Micro-Tom’ using Illumina Hiseq 2500 or Hiseq X Ten system (DDBJ accession No. DRR184874-DRR184876). Together with publicly available NGS data of independent wild-type ‘Micro-Tom’ (TOMJPF00001) (Kobayashi et al. 2014; Pulungan et al. 2018), a total of 10 lines of wild-type NGS data were used as controls to subtract intra-cultivar genomic variations from the DNA variant dataset of 95 mutants as described below.
Bioinformatics analysis of sequence data and acquisition of DNA variant dataset
Short read alignment and subsequent genotype analysis were performed by bowtie2-Genome Analysis Tool Kit (GATK) pipeline. First, artificial sequence of Illumina adaptor was removed from short read data. Then, those with low quality bases were further removed by Fastxtoolkit (https://github.com/agordon/fastx_toolkit) using threshold parameters of Q≥20, p≥90. The resultant NGS reads were aligned to the tomato genome reference SL3.0 with bowtie2 software (Langmead and Salzberg 2012) using the following parameter: “–maxins 500–dovetail–end-to-end–very-sensitive–score-min L, 0, −0.16–mp 2,2–np 1–rdg 1,1–rfg 1,1”. After file format manipulation with Samtools (Li et al. 2009) and Picard tools (https://broadinstitute.github.io/picard/), genotype data were obtained in each sample as genomic VCF (gVCF) with GATK software (McKenna et al. 2010) using the following parameter: “-allowPotentiallyMisencodedQuals -T HaplotypeCaller -ERC GVCF -variant_index_type LINEAR -mmq 5 -forceActive -stand_call_conf 10 -stand_emit_conf 10”. In addition to the exome sequencing dataset obtained in this study (95 mutant lines and one wild-type control), three lines of wild-type exome sequence data and six lines of wild-type whole genome NGS data were also analyzed by the same method to obtain their gVCF. Then, gVCF of 10 wild-type ‘Micro-Tom’ and 95 mutant individuals were combined with the ‘CombineGVCFs’ function of GATK software. Intra-cultivar genomic variations that were present between wild-type ‘Micro-Tom’ individuals were removed from the combined gVCF dataset, and the DNA variants that were present in mutant population with GQ value ≥5 were selected using custom Perl scripts. Frequencies of each DNA variant among 95 mutant lines were also calculated based on the selected variant dataset. The effect of each DNA variant on the protein amino acid sequences was analyzed based on the general feature format (GFF) information of the tomato genome reference Heinz1706 SL3.0/ITAG3.10 using custom Perl scripts.
Results and discussion
Ninety-six plex exome sequencing of ‘Micro-Tom’ mutants
To reveal the frequency and distribution of mutations in the ‘Micro-Tom’ mutants on a population scale, we performed a 96-plex exome capture experiment by modifying a standard protocol of Roche 24-plex SeqCapEz® method. By using dual index 96-plex adaptors and newly designed custom HE oligos, we could obtain exome sequencing data in 95 mutants with an averaged read count of 5.6 million (Supplementary Figure S1a). According to bowtie2 alignment, these short reads could be aligned on the tomato genome reference Heinz1706 SL3.0 at an averaged ratio of 97.0% (Supplementary Figure S1b). In addition, nearly all of the exome capture target regions (49.5 Mb in total length) were covered by the aligned short reads with an averaged read depth of 12.8 (Supplementary Figure S1c, d). Although amounts of aligned reads, covered genomic regions, and read depths were different between samples, it was evident that 96-plex exome sequencing was successfully conducted in this study.
Identification of mutant-specific DNA variant using wild-type controls
Although wild-type ‘Micro-Tom’ strain ‘TOMJPF00001’ that was used for TILLING mutant library development in University of Tsukuba had been subjected to several generations of self-fertilization prior to mutagenesis, it has been suspected that the original ‘TOMJPF00001’ strain was not genetically fixed. Thus, there was a possibility that DNA variant data obtained by exome sequencing in this study contained a large number of intra-cultivar variations that are not caused by mutagenesis treatment but are naturally occurring between wild-type individuals. They have to be computationally removed to analyze DNA variants that are present only in the mutant population. In this study, we used up to 10 lines of wild-type control NGS data (six from whole genome Illumina NGS and four from exome sequencing) to subtract such inter-cultivar DNA variants. The number of DNA variants present in 95 mutants was clearly decreased by increasing the amount of control wild-type data (Table 1). In particular, the number of homozygous mutations was greatly reduced when we used more than nine wild-type controls. When 10 controls were used, 243,531 genotypes were determined as heterozygous variants while 13,479 were determined as homozygous (GQ threshold ≥5). This result was consistent with the fact that genomic DNA samples were bulked between siblings in most mutant lines that were used for exome sequencing. This result suggested that at least several independent lines of wild-type controls are required to subtract intra-cultivar variations and to analyze DNA variants specific to ‘Micro-Tom’ mutants. It was also likely that the original wild-type ‘Micro-Tom’ (TOMJPF00001) was not inbred despite the six generations of self-fertilization. Additional rounds of self-fertilization or creation of double haploid generation may be required to obtain inbred wild-type ‘Micro-Tom.’
| Num of WT controls | GQ threshold | Total count of genotyped allele in 95 mutants | Non-redundant count of identified mutation | DRA accession No. of WT controls | ||
|---|---|---|---|---|---|---|
| REF homozygous | ALT heterozygous | ALT homozygous | ||||
| 1 (a) | 5 | 20,115,830 | 449,828 | 1,608,936 | 330,973 | DRR184873 |
| 4 (b) | 5 | 19,408,161 | 336,001 | 552,310 | 303,293 | (a) + DRR097500, DRR097501, DRR097502 |
| 7 (c) | 5 | 19,229,568 | 297,440 | 251,713 | 263,021 | (b) + DRR097503, DRR097504, DRR000741 |
| 9 (d) | 5 | 18,262,649 | 244,584 | 14,469 | 241,832 | (c) + DRR184874, DRR184875 |
| 10 (e) | 5 | 18,243,610 | 243,531 | 13,479 | 241,391 | (d) + DRR184876 |
| 10 | 10 | 15,635,762 | 239,353 | 9,721 | 235,708 | (e) |
| 10 | 20 | 9,478,983 | 217,775 | 5,757 | 213,883 | (e) |
| 10 | 30 | 3,161,106 | 125,713 | 3,752 | 122,699 | (e) |
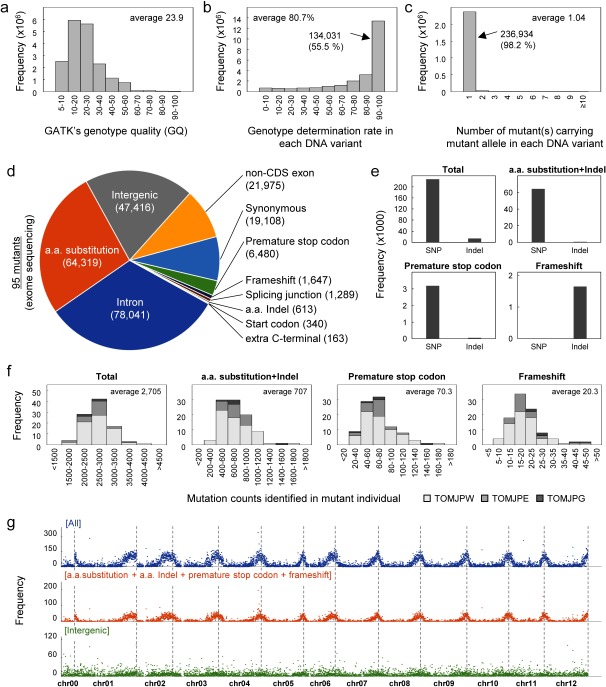
The total non-redundant count of mutations identified in the mutant population was 241,391 under the condition of GQ threshold ≥5 (Table 1). Although this value was greatly reduced under a more stringent condition (e.g., 122,699 in GQ threshold ≥30), we took an approach to include the DNA variant information with low GQ values (GQ threshold ≥5) as a low GQ value itself does not completely reject the reliability of the information. Nevertheless, more than half of the 241,391 DNA variants identified in the mutant population had GQ values of more than 20 (average 23.9, Figure 2a). According to the genotype matrix dataset, around 55.5% of 241,391 DNA variants were genotyped at >90% in the 95 mutants (134,031 variants; Figure 2b). This again demonstrated that our 96-plex exome sequencing method was enough to obtain a mutation dataset on a population scale. Interestingly, 98.2% of 241,391 DNA variants were found to be uniquely present in a single mutant in the population of 95 mutants (Figure 2c). Although such mutation coincidence statistics should be assessed in a larger population (e.g., >10,000 mutants), it was likely that mutations are randomly generated at different genomic positions in the mutant population.
Figure 2. Statistic summary of DNA variants identified in 95 ‘Micro-Tom’ mutants. (a–c) A histogram of (a) GATK’s genotype quality, (b) genotype determination rate, and (c) mutant allele coincidence count in the DNA variant dataset of the 95 ‘Micro-Tom’ mutants. (d) A diagram showing the frequency of mutation type in the DNA variant dataset of 95 mutants. (e) Frequencies of single nucleotide polymorphisms (SNPs) and Indels in each mutation type in the DNA variant dataset of 95 mutants. (f) A histogram of mutation counts that were identified in each mutant individual. (g) Genome-wide distribution and frequency of mutations in the 95 mutants.
Distribution and frequency of mutations that affect protein amino acid sequence in the ‘Micro-Tom’ mutants
Then, we analyzed the effect of each DNA variant on the protein amino acid sequence. In the dataset of 95 mutants, at most 64,319, 6,480 and 1,647 mutations were considered to cause single amino acid substitution, premature stop codon, or frameshift of protein amino acid sequence, respectively (Figure 2d). Most of amino acid substitution mutations and premature stop codon mutations were caused by SNPs while frameshift mutations were caused by Indel (Figure 2e). Although exome capture treatment was conducted before Illumina sequencing, a substantial amount of intergenic and intron mutations was also identified (47,416 or 78,041 in Figure 2d). It is probably because some captured DNA fragment contained the boundary region of exon and intergenic or that of exon and intron. In addition to the exome sequencing dataset, we also analyzed the proportion of amino acid-changing mutations in the DNA variant dataset of 10 wild-type ‘Micro-Tom’ (Supplementary Figure S2a, b). In wild types, at most 25,794, 772, or 951 were considered to cause single amino acid substitution, premature stop codon, and frameshift, respectively, relative to the tomato genome reference Heinz1706 SL3.0/ITAG3.10. Because subtraction effect of intra-cultivar variations was clearly saturated when we used ≥9 wild-type controls (Table 1), the number of wild-type DNA variants shown in Supplementary Figure S2 is unlikely to increase even though additional NGS data of wild-type individuals are added to the analysis. Thus, it is considered that the number of mutations that affect protein amino acid sequence are much higher in mutant populations than the wild-type population.
The averaged counts of mutations present in each mutant were calculated to be 2,705 at most (Figure 2f; GQ threshold ≥5). Those of amino acid substitution mutations (both single substitution and Indel), premature stop codon mutations, and frameshift mutations were calculated to be 707, 70.3, and 20.3, respectively. These values decreased under more stringent GQ threshold condition (e.g., average 1,362 mutations with GQ threshold ≥30). Considering that exome capture was conducted prior to sequencing, the number of mutations present in each mutant must be much higher than this value. Frequency and distribution of mutations were similar between chromosomes in the mutant population unlike wild types (compare Figure 2c and Supplementary Figure S2c), suggesting again that mutations are randomly generated at different genomic positions. Because most of the target region in exome capture were genic regions and genes are abundant at both ends of the chromosome rather than centromeric part, mutation frequencies were high at the end part of the chromosome. Although gamma-ray irradiation is thought to cause large Indels on the genome relative to EMS treatment, frequency of frameshift mutations that were predominantly caused by Indels were clearly not higher in the ‘TOMJPG’ lines (gamma-ray induced mutants) compared with EMS mutants (e.g., ‘TOMJPW’ and ‘TOMJPE’). In contrast to the mutant population, frequency and distribution of DNA variants were vastly different between chromosomes in the dataset of the wild-type population (Supplementary Figure S2c). In particular, chromosome 2, 5, 11 seemed to carry a higher number of DNA variants than other chromosomes against the tomato genome reference Heinz1706 SL3.0.
Based on the population-scale exome mutation dataset obtained in this study, it is possible to conduct in silico screening of a specific mutant that carries a mutation in a gene of interest. One of the sequenced mutants ‘TOMJPW601’ had been previously shown to exhibit a curly leaf phenotype, which is caused by premature stop codon mutation in Solyc09g014380 (SlLAX1) (Pulungan et al. 2018). Because the predicted SlLAX1 protein sequence in the tomato genome reference Heinz1706 SL3.0/ITAG3.10 has additional 76 amino acids at the N-terminal relative to that of SL2.5, this mutations is equivalent to W261* in SL3.0/ITAG3.10 while W185* in SL2.5. In this study, our 96-plex exome sequencing successfully identified the W261* mutation, indicating that the exome mutation dataset can be used for in silico mutant screening (Table 2). Mutations could be also found in other genes such as essential regulator of fruit ripening; CNR, NOR, and RIN (Barry et al. 2000; DellaPenna et al. 1989; Giovannoni 2004; Lincoln and Fischer 1988; Orfila et al. 2002; Supplementary Table S3). In total, 22,353 genes were found to have some mutations that affect protein amino acid sequences in the mutation dataset of 95 mutants. The mutation data file is provided as supplementary data 1 (compressed tab-delimited text). It contains information on mutations (DNA variants), their effects on protein amino acid sequence, as well as ‘TOMJP’ ID information of mutant(s) that carry the corresponding mutation. It is possible to conduct in silico screening on a Linux server by typing the command like “zcat supplementary_data_1.tsv.gz | grep Solyc09g014380 > list_of_mutants_ Solyc09g014380.txt”.
| mutant ID | DP | GQ | Chr. | Position (bp) | REF | ALT | location | mutation type | aa change |
|---|---|---|---|---|---|---|---|---|---|
| Solyc09g014380 (SlLAX1) | |||||||||
| TOMJPE5066_1 | 6 | 31 | SL3.0ch09 | 6008688 | G | T | CDS.1 | missense | D32Y |
| TOMJPE6034_1 | 6 | 43 | SL3.0ch09 | 6008839 | AT | A | intron.1 | intron | — |
| TOMJPW1163_1 | 6 | 31 | SL3.0ch09 | 6008853 | G | T | intron.1 | intron | — |
| TOMJPE5262_1 | 6 | 31 | SL3.0ch09 | 6008954 | C | A | CDS.2 | missense | T94N |
| TOMJPW601_1 | 5 | 35 | SL3.0ch09 | 6009537 | G | T | intron.4 | intron | — |
| TOMJPW499_1 | 5 | 67 | SL3.0ch09 | 6009575 | G | A | intron.4 | intron | — |
| TOMJPW499_1 | 5 | 67 | SL3.0ch09 | 6009577 | T | G | intron.4 | intron | — |
| TOMJPW601_1 | 10 | 30 | SL3.0ch09 | 6010742 | G | A | CDS.6 | nonsense | W261* |
| TOMJPW1160_1 | 15 | 54 | SL3.0ch09 | 6011197 | T | C | intron.6 | intron | — |
Conclusion
In this study, we demonstrated that 96-plex exome sequencing is an efficient method to identify genome-wide mutations in the ‘Micro-Tom’ TILLING population. We also demonstrated that several wild-type controls are required to extract the DNA variants (mutations) that are specifically present in the mutant population. Taken together, these attempts successfully revealed genome-wide distribution and frequency of mutations in the ‘Micro-Tom’ mutants on a population scale. Because the sequence throughput of NGS instrumentation (e.g., Illumina Novaseq®) is improving, it may be possible to conduct larger scale multiplex sequencing in the future (e.g., 384-plex analysis). Such high-throughput genotyping methods may open a way to construct a more comprehensive exome mutation dataset in tens of thousands of ‘Micro-Tom’ mutants, which in turn enables us to conduct in silico screening of specific tomato mutants in a more comprehensive manner. Together with other breeding technology, it will contribute to both basic research of gene function and breeding of new cultivars.
Acknowledgments
We thank Ms. Y. Fujimori, N. Inage, R. Masuda, M. Miyamoto, and M. Yamaguchi for their skilled technical assistance. This work was supported by JSPS KAKENHI Grant-in-Aid for Research Activity Start-up to R.Y. (Grant No. 15H06071), JSPS KAKENHI Grant-in-Aid for Young Scientists (B) to Y.O. (Grant No. 16K18633), and the National BioResource Project (NBRP) of the Japan Agency for Research and Development (AMED) to K.H. and H.E. (Grant No. 18km0210110j0002).
Abbreviations
- NGS
Next-generation sequencing
- TILLING
Targeting Induced Local Lesions in Genomes
- SNPs
single nucleotide polymorphisms
- EMS
ethyl methanesulfonate
- VCF
variant call format
Author contributions
The authors primarily contributed to this work are R.Y., K.H., and H.E. R.Y. and H.E. supervised the entire part of this project. Bioinformatics analysis and exome library construction were performed by R.Y. Plant cultivation and genomic DNA extraction in 96 ‘Micro-Tom’ mutants were performed by K.H. NGS library construction and NGS dataset acquisitions were also contributed by Y.O., N.W., P.T.D., P.S.I., H.S., and T.A.
Accession Numbers
Nucleotide sequence data reported in this study are available in the DDBJ/EMBL/GenBank databases under the following accession numbers: DRR184778-DRR184876.
Disclosure Statement
All authors have no conflict of interest.
Supplementary Data
References
- Aoki K, Yano K, Suzuki A, Kawamura S, Sakurai N, Suda K, Kurabayashi A, Suzuki T, Tsugane T, Watanabe M, et al. (2010) Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genomics 11: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldet P, Bres C, Okabe Y, Mauxion JP, Just D, Bournonville C, Ferrand C, Mori K, Ezura H, Rothan C (2013) Investigating the role of vitamin C in tomato through TILLING identification of ascorbate-deficient tomato mutants. Plant Biotechnol 30: 309–314 [Google Scholar]
- Baker M (2012) De novo genome assembly: What every biologist should know. Nat Methods 9: 333–337 [Google Scholar]
- Barry CS, Llop-Tous MI, Grierson D (2000) The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol 123: 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, Lincoln JE, Fischer RL, Bennett AB (1989) Transcriptional analysis of polygalacturonase and other ripening associated genes in Rutgers, rin, nor, and Nr tomato fruit. Plant Physiol 90: 1372–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Vincent K, Sharp P (2009) Simultaneous mutation detection of three homoeologous genes in wheat by High Resolution Melting analysis and Mutation Surveyor®. BMC Plant Biol 9: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliot F, Cordeiro G, Bundock PC, Henry RJ (2008) SNP discovery by ECOTILLING using capillary electrophoresis. Plant Genotyping 2: 78–87 [Google Scholar]
- Gady AL, Hermans FW, Van de Wal MH, Van Loo EN, Visser RG, Bachem CW (2009) Implementation of two high through-put techniques in a novel application: detecting point mutations in large EMS mutated plant populations. Plant Methods 5: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauffier C, Lebaron C, Moretti A, Constant C, Moquet F, Bonnet G, Caranta C, Gallois JL (2016) A TILLING approach to generate broad-spectrum resistance to potyviruses in tomato is hampered by eIF4E gene redundancy. Plant J 85: 717–729 [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ (2004) Genetic regulation of fruit development and ripening. Plant Cell 16(Suppl): S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene EA, Codomo CA, Taylor NE, Henikoff JG, Till BJ, Reynolds SH, Enns LC, Burtner C, Johnson JE, Odden AR, et al. (2003) Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Reddaiah B, Salava H, Upadhyaya P, Tyagi K, Sarma S, Datta S, Malhotra B, Thomas S, Sunkum A, et al. (2017) Next-generation sequencing (NGS)-based identification of induced mutations in a doubly mutagenized tomato (Solanum lycopersicum) population. Plant J 92: 495–508 [DOI] [PubMed] [Google Scholar]
- Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Toki S (2015) CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem Biophys Res Commun 467: 76–82 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Nagasaki H, Garcia V, Just D, Bres C, Mauxion JP, Le Paslier MC, Brunel D, Suda K, Minakuchi Y, et al. (2014) Genome-wide analysis of intraspecific DNA polymorphism in ‘Micro-Tom,’ a model cultivar of tomato (Solanum lycopersicum). Plant Cell Physiol 55: 445–454 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln JE, Fischer RL (1988) Regulation of gene expression by ethylene in wild-type and rin tomato (Lycopersicon esculentum) fruit. Plant Physiol 88: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochlainn SÓ, Amoah S, Graham NS, Alamer K, Rios JJ, Kurup S, Stoute A, Hammond JP, Østergaard L, King GJ, et al. (2011) High Resolution Melt (HRM) analysis is an efficient tool to genotype EMS mutants in complex crop genomes. Plant Methods 7: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura C, Yamaguchi I, Inamura M, Ban Y, Kobayashi Y, Yin Y, Saito T, Kuwata C, Imanishi S, Nishimura S (2007) Generation of gamma irradiation-induced mutant lines of miniature tomato (Solanum lycopersicum L.). Plant Biotechnol 24: 39–44 [Google Scholar]
- Mazzucato A, Cellini F, Bouzayen M, Zouine M, Mila I, Minoia S, Petrozza A, Picarella ME, Ruiu F, Carriero F (2015) A TILLING allele of the tomato Aux/IAA9 gene offers new insights into fruit set mechanisms and perspectives for breeding seedless tomatoes. Mol Breed 35: 22 [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010) The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner R, Jacobson Y, Melamed S, Levyatuv S, Shalev G, Ashri A, Elkind Y, Levy A (1997) A new model system for tomato genetics. Plant J 12: 1465–1472 [Google Scholar]
- Minoia S, Petrozza A, D’Onofrio O, Piron F, Mosca G, Sozio G, Cellini F, Bendahmane A, Carriero F (2010) A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Research Notes 3: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves LG, Davis JM, Barbazuk WB, Kirst M (2013) Whole-exome targeted sequencing of the uncharacterized pine genome. Plant J 75: 146–156 [DOI] [PubMed] [Google Scholar]
- Okabe Y, Asamizu E, Saito T, Matsukura C, Ariizumi T, Brès C, Rothan C, Mizoguchi T, Ezura H (2011) Tomato TILLING technology: Development of a reverse genetics tool for the efficient isolation of mutants from Micro-Tom mutant libraries. Plant Cell Physiol 52: 1994–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y, Ariizumi T, Ezura H (2013) Updating the micro-tom TILLING platform. Breed Sci 63: 42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orfila C, Huisman MM, Willats WG, van Alebeek GJW, Schols HA, Seymour GB, Knox PJ (2002) Altered cell wall disassembly during ripening of Cnr tomato fruit: Implications for cell adhesion and fruit softening. Planta 215: 440–447 [DOI] [PubMed] [Google Scholar]
- Piron F, Nicolaï M, Minoïa S, Piednoir E, Moretti A, Salgues A, Zamir D, Caranta C, Bendahmane A (2010) An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS One 5: e11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulungan SI, Yano R, Okabe Y, Ichino T, Kojima M, Takebayashi Y, Sakakibara H, Ariizumi T, Ezura H (2018) SlLAX1 is required for normal leaf development mediated by balanced adaxial and abaxial pavement cell growth in tomato. Plant Cell Physiol 59: 1170–1186 [DOI] [PubMed] [Google Scholar]
- Rigola D, van Oeveren J, Janssen A, Bonné A, Schneiders H, van der Poel HJ, van Orsouw NJ, Hogers RC, de Both MT, van Eijk MJ (2009) High-throughput detection of induced mutations and natural variation using KeyPoint™ technology. PLoS One 4: e4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Ariizumi T, Okabe Y, Asamizu E, Hiwasa-Tanase K, Fukuda N, Mizoguchi T, Yamazaki Y, Aoki K, Ezura H (2011) TOMATOMA: A novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol 52: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglione D, Pinosio S, Marroni F, Di Centa E, Fornasiero A, Magris G, Scalabrin S, Cattonaro F, Taylor G, Morgante M (2019) Single primer enrichment technology as a tool for massive genotyping: A benchmark on black poplar and maize. Ann Bot 124: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Harbaugh BK (1989) Micro-Tom: A miniature dwarf tomato. Agricultural Experiment Station, Institute of Food and Agricultural Sciences, University of Florida, Circular, S-370: 1–6 [Google Scholar]
- Shikata M, Hoshikawa K, Ariizumi T, Fukuda N, Yamazaki Y, Ezura H (2015) TOMATOMA update: Phenotypic and metabolite information in the Micro-Tom mutant resource. Plant Cell Physiol 57: e11. [DOI] [PubMed] [Google Scholar]
- Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, et al. (2017) Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol 35: 441–443 [DOI] [PubMed] [Google Scholar]
- Shirasawa K, Hirakawa H, Nunome T, Tabata S, Isobe S (2016) Genome-wide survey of artificial mutations induced by ethyl methanesulfonate and gamma rays in tomato. Plant Biotechnol J 14: 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HJ, Uchii S, Watanabe S, Ezura H (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol 47: 426–431 [DOI] [PubMed] [Google Scholar]
- Takagi K, Yano R, Tochigi S, Fujisawa Y, Tsuchinaga H, Takahashi Y, Takada Y, Kaga A, Anai T, Tsukamoto C, et al. (2018) Genetic and functional characterization of Sg-4 glycosyltransferase involved in the formation of sugar chain structure at the C-3 position of soybean saponins. Phytochemistry 156: 96–105 [DOI] [PubMed] [Google Scholar]
- Takayama M, Matsukura C, Ariizumi T, Ezura H (2017) Activating glutamate decarboxylase activity by removing the autoinhibitory domain leads to hyper γ-aminobutyric acid (GABA) accumulation in tomato fruit. Plant Cell Rep 36: 103–116 [DOI] [PubMed] [Google Scholar]
- The Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Kaga A, Anai T, Shimizu T, Sayama T, Takagi K, Machita K, Watanabe S, Nishimura M, Yamada N, et al. (2015) Construction of a high-density mutant library in soybean and development of a mutant retrieval method using amplicon sequencing. BMC Genomics 16: 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-Mas R, Bea S, Puente DA, López-Otín C, Puente XS (2012) Estimation of copy number alterations from exome sequencing data. PLoS One 7: e51422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Mizoguchi T, Aoki K, Kubo Y, Mori H, Imanishi S, Yamazaki Y, Shibata D, Ezura H (2007) Ethylmethansulfonate (EMS) mutagenesis of Solanum lycopersicum cv. Micro-Tom for large-scale mutant screens. Plant Biotechnol 24: 33–38 [Google Scholar]
- Yano R, Takagi K, Takada Y, Mukaiyama K, Tsukamoto C, Sayama T, Kaga A, Anai T, Sawai S, Ohyama K, et al. (2017) Metabolic switching of astringent and beneficial triterpenoid saponins in soybean is achieved by a loss-of-function mutation in cytochrome P450 72A69. Plant J 89: 527–539 [DOI] [PubMed] [Google Scholar]
- Yano R, Takagi K, Tochigi S, Fujisawa Y, Nomura Y, Tsuchinaga H, Takahashi Y, Takada Y, Kaga A, Anai T, et al. (2018) Isolation and characterization of the soybean Sg-3 gene that is involved in genetic variation in sugar chain composition at the C-3 position in soyasaponins. Plant Cell Physiol 59: 792–810 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Smith SM, Ayele M, Yang L, Jogi A, Chaluvadi SR, Bennetzen JL (2012) High-throughput discovery of mutations in tef semi-dwarfing genes by next-generation sequencing analysis. Genetics 192: 819–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.