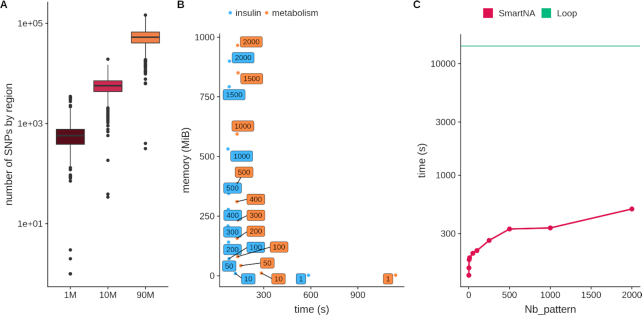
Figure 3.
Resource usage and computation time. (A) The distribution of the number of SNPs per region when using the recombination hotspot map proposed by Berisa and Pickrell (27) while using 1M SNPs from the Illumina Omni1-QuAd, 9.9M SNPs from a recent GWAS and 96M SNPs as available in the latest UK Biobank genotype panel after imputation. (B) The trade-off between memory usage and computation time. JASS omnibus statistics was computed for 4 traits and 3 192 045 SNPs (insulin) and 11 traits and 3 673 285 SNPs (metabolism). The y-axis is the peak in memory usage in megabytes and x-axis is the time (in seconds) to perform the task. Labels attached to data points give the number of genomic regions processed at once. For example the two boxes filled with a “2000" in the top left of the plot correspond to the case were the entire dataset is loaded in memory (as there is a total of 1703 regions in this example). Color represents the group of traits. (C) The computation time optimization. We computed the omnibus statistics for 3 673 285 SNPs for 11 traits; the y-axis is the time in seconds (logarithmic scale) and x-axis is the number of missing data in a pattern. The straight green line shows the result from a naïve computation loop approach over all SNPs, and the red line shows the results from our optimized algorithm.