Figure 4.
BFD1 Is Required to Express Bradyzoite Genes and Initiate Differentiation
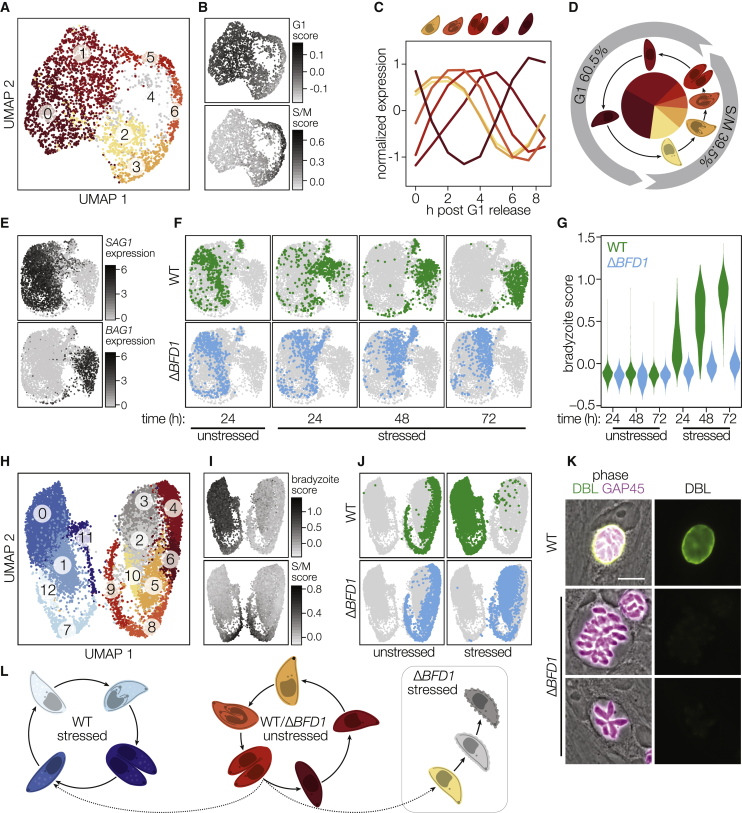
(A) Clustering of unstressed parasites of both genotypes after 72 h of growth (3,149 cells total) visualized by uniform manifold approximation and projection (UMAP).
(B) UMAP from (A) shaded by score for expression of known G1 or S/M-specific gene sets.
(C) Average expression profiles of all genes differentially expressed within each cluster identified in (A) across a microarray dataset of synchronized tachyzoites (Behnke et al., 2010). Colors correspond to those used in (A).
(D) Proportion of cells in G1 (0 and 1) or S/M (2, 3, 5, and 6) clusters.
(E) Cells shaded by expression of the canonical stage-specific genes SAG1 and BAG1 following clustering of all parasites from all time points, genotypes, and growth conditions. UMAP visualization is downsampled to 500 cells from each combination of time point, genotype, and growth condition (6,000 cells total).
(F) UMAP as in (E), with cells highlighted by sample of origin for WT (green) or ΔBFD1 (blue) parasites.
(G) Distribution of cell scores for the expression of genes highly upregulated in bradyzoites.
(H) UMAP of all WT and ΔBFD1 parasites from unstressed and stressed cultures at 72 h time point.
(I and J) UMAP projection from (H), shaded by scores for expression of highly upregulated bradyzoite genes as in G, or S/M-specific genes as in (B) (I) or colored by sample of origin (J).
(K) Representative WT and ΔBFD1 vacuoles at 72 h post-alkaline stress. GAP45 is a marker for the inner membrane complex. Scale bar is 10 μm.
(L) Proposed model of cell-cycle progression for WT and ΔBFD1 parasites under the different treatments.