Figure 1.
FAMIN Is a Purine-Nucleoside-Metabolizing Enzyme
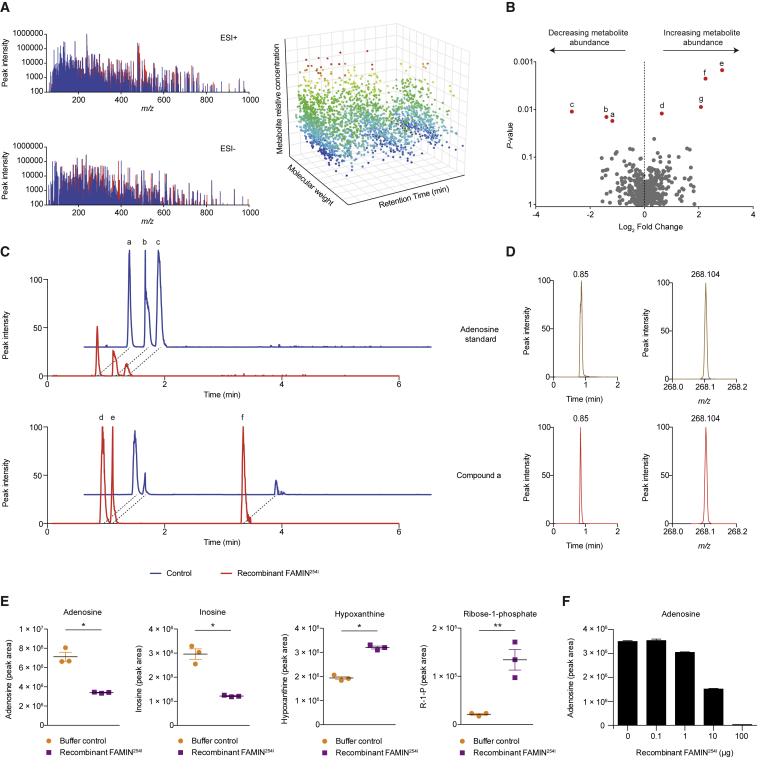
(A) Metabolomic library of HepG2 cells after transfection with FAMIN siRNA. Representative total mass spectra (left) separated by molecular weight (m/z), chromatography retention time, and relative levels (right).
(B) Change in relative metabolite levels in the library after incubation with recombinant FAMIN254I or protein buffer control, depicted as volcano plot with unadjusted p values. Red dots, candidate substrates and products whose abundance decreased (a–c) or increased (d–f; n = 3 independent reactions).
(C) Representative extracted chromatograms for candidate substrates (top) and products (bottom) by using normalized peak intensity for each given m/z value.
(D) Representative mass spectra and extracted chromatograms for compound a and corresponding authentic standard.
(E) Levels of adenosine, inosine, hypoxanthine, and ribose-1-phosphate (R1P) within the metabolomic library incubated with FAMIN254I or protein buffer control (n = 3, mean ± SEM).
(F) Levels of adenosine within the metabolomic library incubated with 0.1–100 μg of FAMIN254I or protein buffer control (n = 3, mean ± SEM).
Data representative of at least 3 independent experiments. ∗p < 0.05 and ∗∗p < 0.01 (unpaired, two-tailed Student’s t test).