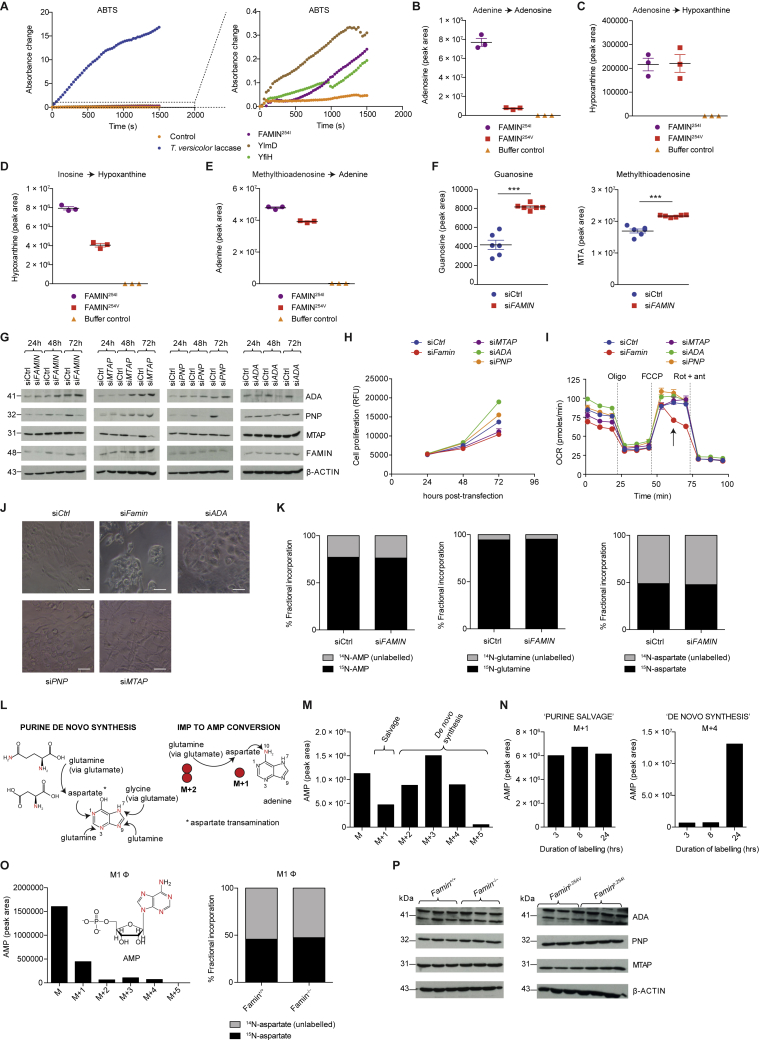
Figure S3.
FAMIN Enzymatic Activity Determines Cellular Purine Metabolism and Is Impaired in FAMIN254V, Related to Figure 3
(A) Laccase enzyme activity of recombinant YlmD, YfiH, FAMIN254I and Trametes versicolor laccase using 2,2′-azino-bis-3-ethylbenzothiazoline-6-sulphonic acid (ABTS). Please note the right panel’s y axis, which shows a graphical enlargement of the low absorbance readings from the left panel. Data are representative of 2-3 independent experiments.
(B) Adenosine levels following incubation of 10 μg Strep-tagged FAMIN254I, FAMIN254V or control with 50 μM adenine and 50 μM ribose-1-phosphate for 1 h in 100 μL PBS.
(C) Hypoxanthine levels following incubation of 10 μg recombinant Strep-tagged FAMIN254I or FAMIN254V with 10 μM adenosine for 1 h in 100 μL phosphate buffered saline (PBS), (n = 3).
(D) EC 2.4.2.1 (purine nucleoside phosphorylase) and (E) EC 2.4.2.28 (MTA phosphorylase) activities of Strep-tagged FAMIN254I and FAMIN254V as measured by hypoxanthine and adenine following incubation of recombinant protein with 10 μM inosine and methylthioadenosine (MTA), respectively, in PBS (n = 3).
(F) Guanosine and S-methyl-5′-thioadenosine levels in control and FAMIN-silenced HepG2 cells 48 h after transfection (n = 6).
(G) Immunoblots (IB) for ADA, PNP, MTAP and FAMIN from HepG2 cell lysates silenced for FAMIN (siFAMIN), adenosine deaminase (siADA), purine nucleoside phosphorylase (siPNP), methylthioadenosine phosphorylase (siMTAP) or scrambled siRNA (siCtrl) at 24 h, 48 h or 72 h following transfection; β-ACTIN loading control.
(H) Cell proliferation over time of control, FAMIN, ADA, PNP or MTAP silenced HepG2 cells as measured by CyQUANT assay (n = 12).
(I) Oxygen consumption rate (OCR) of HepG2 cells transfected with siFAMIN, siADA, siPNP, siMTAP or siCtrl. Basal OCR measurement was followed by sequential treatment (dotted vertical lines) with oligomycin A (Oligo), FCCP, and rotenone plus antimycin A (Rot + ant), (n = 3). Arrow indicates steep decline observed in siFAMIN cells after treatment with FCCP.
(J) Cell morphology by light microscopy of HepG2 cells silenced with siFAMIN, siADA, siPNP, siMTAP or siCtrl as observed at 72 h after transfection; scale bar = 125 μm. Data are representative of at least 3 independent experiments.
(K–O) A 24h pulse with 15N2-glutamine labeled three quarters of AMP in HepG2 cells (n = 3; mean). Since this pulse labeled almost all cellular glutamine and ∼50% of aspartate, the number of incorporated 15N atoms into AMP allowed (L–M) estimating the proportion of purine de novo synthesis (M+2, M+3, M+4 isotopomers) versus salvage via HPRT (M+1 isotopomer). (N) HepG2 cells engaged both purine salvage and de novo synthesis, but as expected with different kinetics. (O) Terminally differentiated M1Φ, in contrast, exhibited very little de novo synthesis, Levels of M, M+1, M+2, M+3, M+4, M+5 labeled forms of AMP in M1 macrophages after a 24 h pulse with 15N2-glutamine (n = 3; mean). The M+1 isotopomer might substantially underestimate salvage, since only half of aspartate is labeled and any HPRT-dependent salvage subsequent to de novo synthesis, or salvage via APRT, would not be captured. This high turnover of the purine ring extends to all essential metabolites and cofactors that contain adenyl groups, i.e., coenzyme A (CoA), acetyl-CoA, flavin adenine dinucleotide (FAD), nicotinamide adenine dinucleotide (NAD), NAD phosphate (NADP), SAM, SAH and MTA (data not shown).
(P) Immunoblots (IB) for adenosine deaminase (ADA), purine nucleoside phosphorylase (PNP) and S-methyl-5′-thioadenosine phosphorylase (MTAP) in Famin+/+ and Famin–/– and Faminp.254V and Faminp.254I M1 macrophages; β-actin, loading control.
Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (unpaired, two-tailed Student’s t test).