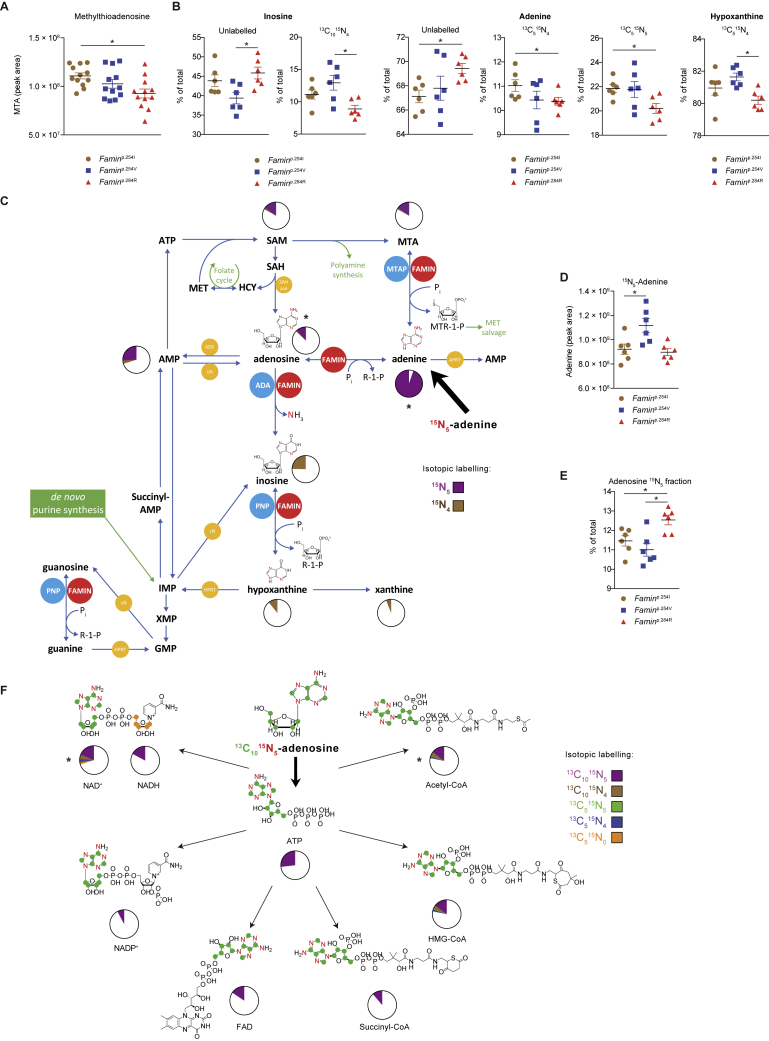
Figure S4.
FAMIN Affects Routing through Central Purine Metabolism, Related to Figure 4
(A) Methylthioadenosine levels in Faminp.254I, Faminp.254V, Faminp.284R M1 macrophages (n = 12).
(B) Fraction of inosine, adenine and hypoxanthine labeled as the indicated isotopomer in M1 macrophages after a 3 h pulse with [13C1015N5] adenosine (n = 6).
(C) Metabolic fate of stable isotope-labeled [15N5] adenine after a 3 h pulse of M1 macrophages (n = 6; mean). Schematic representation of central purine metabolism. AMP can be generated from adenine via sequential FAMIN and ADK activities or via APRT without loss of label (purple). Labeled [15N5] adenosine can be deaminated into inosine by FAMIN or ADA, with a loss of a single 15N as ammonia, generating a [15N4] isotopomer (brown). Routes of interconversion and relationship with other metabolic pathways are also illustrated. Fractions of different labeled states averaged across genotypes following the 3 h pulse with [15N5] adenine are depicted as pie charts; asterisks indicate metabolites with significantly altered isotopic labeling across genotypes as depicted in Figures S4D and S4E. Adenine phosphoribosyl transferase (APRT); adenosine kinase (ADK); adenosine deaminase (ADA); cytosolic nucleotidase (cN); hypoxanthine-guanine phosphoribosyl transferase (HPRT); purine nucleoside phosphorylase (PNP); S-methyl-5′-thioadenosine phosphorylase (MTAP); S-adenosylhomocysteine hydrolase (SAHase).
(D) [15N5] adenine levels in M1 macrophages after a 3 h pulse with [15N5] adenine (n = 6).
(E) Fraction of adenosine labeled as the [15N5] isotopomer in M1 macrophages after a 3 h pulse with [15N5] adenine (n = 6).
(F) Fraction of ATP, NAD+, NADH, FAD, acetyl-CoA, HMG-CoA and succinyl-CoA labeled as the indicated isotopomer in M1 macrophages after a 3 h pulse with [13C1015N5] adenosine. Fractions of different labeled states (averaged across Faminp.254I, Faminp.254V and Faminp.284R genotypes) are depicted as pie charts. Asterisks indicate significantly altered isotopic labeling across Famin genotypes (n = 6 per genotype).
Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (unpaired, two-tailed Student’s t test or one-way ANOVA).