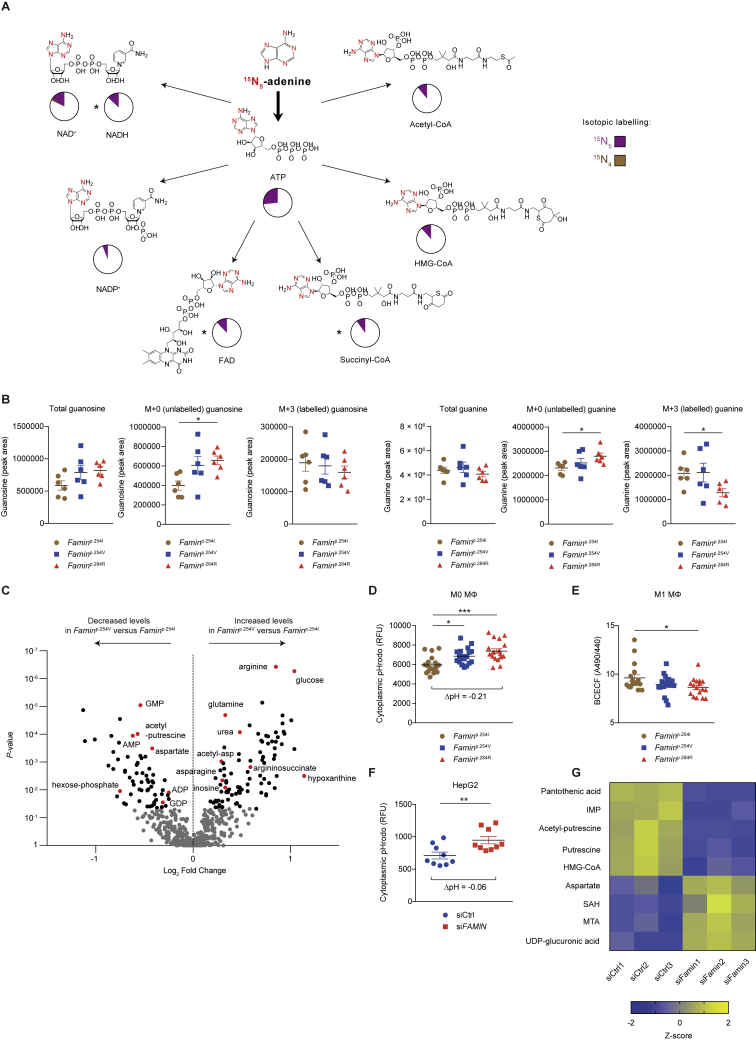
Figure S5.
FAMIN Activity Affects Purine-Containing Cofactor Turnover, Related to Figure 4
(A) Fraction of ATP, NAD+, NADH, FAD, acetyl-CoA, HMG-CoA and succinyl-CoA labeled as the indicated isotopomer in M1 macrophages after a 3 h pulse with [15N5] adenine. Fractions of different labeled states (averaged across Faminp.254I, Faminp.254V and Faminp.284R genotypes) are depicted as pie charts. Asterisks indicate significantly altered isotopic labeling across Famin genotypes (n = 6 per genotype).
(B) Total, unlabelled and [13C115N2] guanine and guanosine levels in M1 macrophages after a 3 h pulse with [13C115N2] guanosine (n = 6).
(C) Differential metabolite levels in Faminp.254I versus Faminp.254V M1 macrophages. Data depicted as a volcano plot using p value and log2 fold change. Grey dots are non-significant, while black dots correspond to LCMS features with significantly altered abundance following Benjamini-Hochberg correction for multiple testing. Differential metabolites shown in red were confirmed and identified as indicated (n = 6).
(D) Cytoplasmic pH measured using pHrodo in Faminp.254I, Faminp.254V, Faminp.284R M0 macrophages (n = 18)
(E) Cytoplasmic pH measured using BCECF with dual excitation at 440nm and 490nm in Faminp.254I, Faminp.254V, Faminp.284R M1 macrophages (n = 18). Reduced 490:440 ratio corresponds to lower (more acidic) pHc.
(F) Cytoplasmic pH measured using pHrodo in control and FAMIN-silenced HepG2 cells 48 h after transfection (n = 9).
(G) Heatmap of metabolites in control and FAMIN-silenced HepG2 cells 24 h after transfection (n = 3).
Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (unpaired, two-tailed Student’s t test or one-way ANOVA).