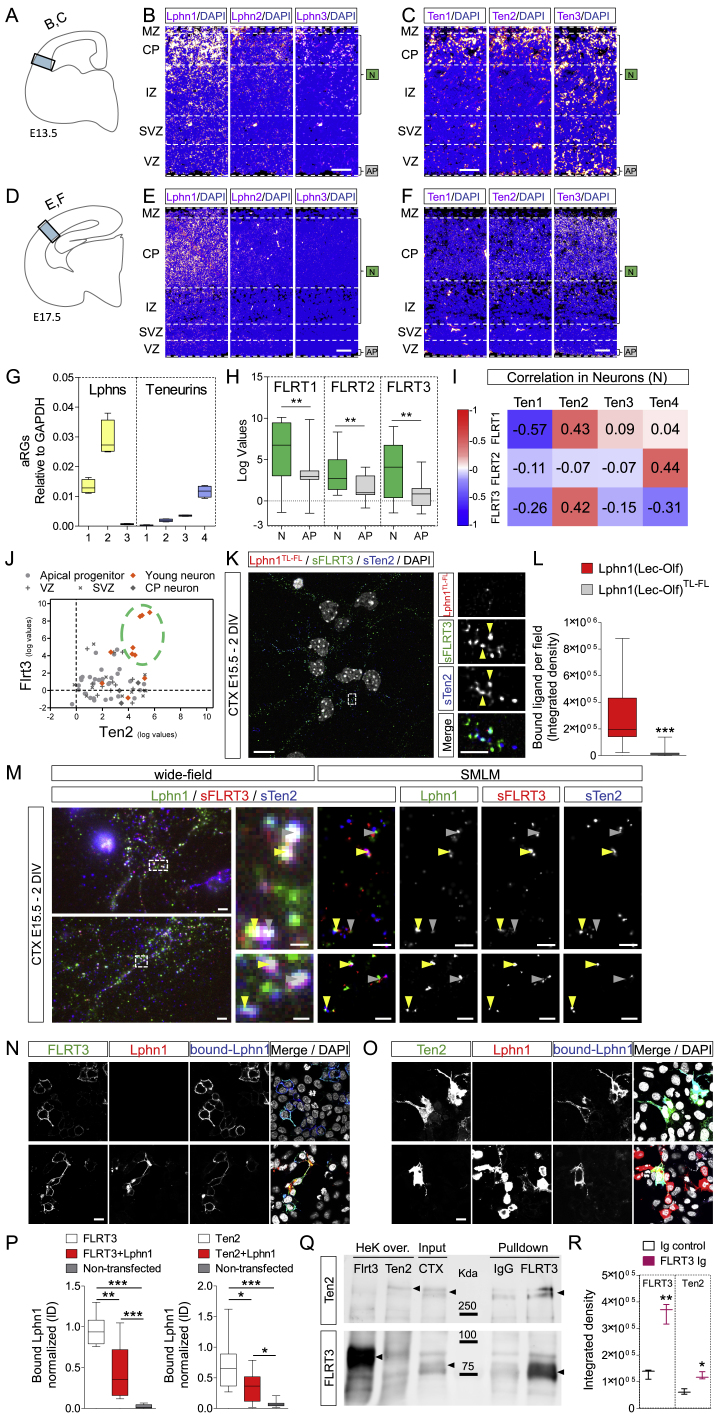
Figure S3.
Teneurins, Latrophilins, and FLRTs Are Expressed during Cortical Development, Related to Figure 3
(A) Diagram showing the location of the coronal cortical region shown on panels B and C. (B and C) ISH for all Latrophilins (B) and Ten1,2 and 3 (C) colored in magenta show protein expression in the cortex of coronal sections of E13.5 mouse embryos. Nuclear staining with DAPI is shown in blue. (D) Diagram showing the location of the cortical region is shown on panels E and F. (E) ISH for all Latrophilins show that Lphn1 is expressed in the cortex of coronal sections of E17.5 mouse embryos. (F) ISH for Ten1, 2 and 3 show higher expression of Ten2 and 3 in the cortex of coronal sections of E17.5 mouse embryos. (G) Latrophilin and Teneurin expression levels normalized to GAPDH in mouse apical RG cells, using RNA profiling data published in Florio et al. (2015) (GSE65000). Lphn2 shows high expression levels compared to Lphn1 and Ten4. The data are presented as whisker plots. (H) FLRT1-3 expression in neurons (N) and apical progenitors (AP) using RNA profiling data published in Kawaguchi et al. (2008) (GEO: GSE10881). FLRT mRNA levels are high in neurons (N) compared to apical progenitors (AP). ∗∗p < 0.01, two-tailed Student’s t test. The data are presented as whisker plots. (I) Correlation analysis using RNA-Seq data for FLRTs and Teneurins in neurons, using data published in GEO: GSE10881. Ten2 expression correlates with FLRT1 and FLRT3, meaning it is present in the same neurons, while Ten4 expression shows correlation with FLRT2. (J) Scatter diagram showing data from 70 single cells from E14.5 mouse cortex, showing the variation in gene expression for FLRT3 and Ten2. The cluster of young neurons shows the strongest expression of both FLRT3 and Ten2. We used raw data previously published in GEO: GSE10881. (K) Surface staining for FLRT3 (red) and Ten2 (green) on E15.5 cortical neurons after 2 days of in vitro culture (DIV) treated with Lphn1TL-FL (Lec-Olf) proteins for 20 min at room temperature. FLRT3 and Ten2 show some degree of co-localization (yellow arrowheads) (see quantification Figure 3I). The area in the dashed rectangle is magnified on the right. (L) Quantification of Lphn1 and the TL-FL double mutant binding to cortical neurons. The double mutant protein shows almost no binding to these cultures. n > 30 fields from 3 experiments. ∗∗∗p < 0.001, two-tailed Student’s t test. (M) Single Molecule Localization Microscopy (SMLM) imaging of E15.5 cortical neurons after 2 days in vitro (DIV), surface stained for surface FLRT3 (red) and surface Ten2 (blue) and treated with Lphn1 protein (green) for 20min at RT. Yellow arrowheads indicated co-localization within the SMLM resolution of approx. 30 nm. Gray arrowheads indicate signals that show co-localization in the conventional wide-field fluorescence, but only close proximity in SMLM. (N) Lphn1 (Lec-Olf), blue, binds to HEK293 cells expressing FLRT3 (green) in the presence or absence of Lphn1 in cis (red). Nuclei are stained with DAPI. (O) Lphn1 (Lec-Olf), blue, binds to HEK293 cells expressing Ten2 (green) in the presence of absence of Lphn1 in cis (red). Nuclei are stained with DAPI. (P) Quantification of data shown in panels (N) and (O). Lphn1 (Lec-Olf) binding was normalized with the intensity of the FLRT3 (N), Ten (O) or non-transfected signal. n > 10 fields. ∗p < 0.05, ∗∗p < 0.01, two-tailed Student’s t test. (Q) Control and FLRT3 pulldowns from mouse cortex E15.5 were analyzed by western blot. On the first two lanes we loaded 40μg of HEK cells overexpressing FLRT3 and Ten2, respectively, as a positive control. The third lane has 60μg of cortical lysate (CTX) input. The last two lanes are Ig control (left) and FLRT3 (right) pulldowns from 1mg CTX tissue. Ten2 and FLRT3 protein bands are indicated with black arrowheads. (R) Quantification of data shown in Q from 3 pull-down experiments (n = 3). ∗p < 0.05, ∗∗p < 0.01, two-tailed Student’s t test. Scale bar = 75 μm (B-F), 15 μm (K, N, O), 2 μm (inset of K, overview images in M), 500 nm (inset of M).