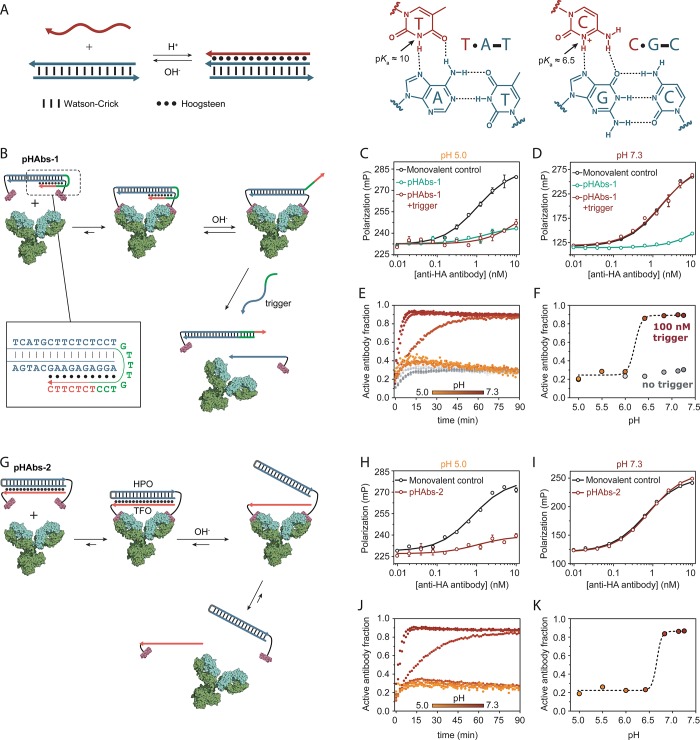
Figure 1.
Antibody activation at high pH. (A) DNA-based triple helices are formed by standard antiparallel Watson–Crick base pairing (dash) and pH-sensitive parallel Hoogsteen interactions (dots). Triple helix formation is governed by protonation of the N3 of thymine (pKa ∼ 10) or cytosine (pKa ∼ 6.5). (B) Design of bivalent antibody ligand pHAbs-1, assembled via a 20 base pair DNA duplex containing a 15 nucleotide overhang that folds back on the duplex to form a 10 nucleotide triple helix. (C, D) Competition assays between a fluorescently labeled HA peptide epitope and pHAbs-1 or a monovalent control in absence and presence of 100 nM trigger oligonucleotide at pH 5.0 and 7.3, respectively. (E) Fraction of activated antibody in time when blocked by pHAbs-1 in absence (gray symbols) and presence of 100 nM trigger oligonucleotide (colored symbols) at various pH values. (F) Steady state fraction of activated antibody at 90 min as a function of pH. (G) Design of bivalent antibody ligand pHAbs-2, assembled through the formation of a 20 nucleotide intermolecular triple helix between an epitope-functionalized DNA hairpin (HPO) and an epitope-functionalized triplex forming oligonucleotide (TFO), each conjugated to a HA-tag peptide epitope. (H, I) Competition assays between a fluorescently labeled HA peptide epitope and pHAbs-2 or a monovalent control at pH 5.0 and 7.3, respectively. (J) Fraction of activated antibody in time when blocked by pHAbs-2 at various pH values. (K) Steady state fraction of activated antibody at 90 min as a function of pH. Error bars represent standard error of the mean of duplicate measurements.