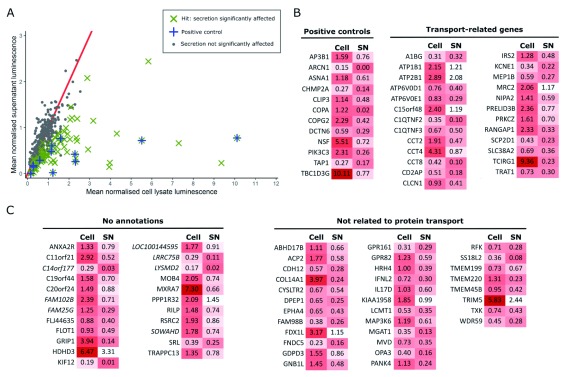
Figure 2. Horseradish peroxidase (HRP) screen to identify genes important for secretion.
Approximately 400 genes were screened in an arrayed siRNA knockdown format. ( A) Mean of the normalised luminescence for both cell lysate and supernatant signals from two independent replicates. ~400 genes were screened; those with significant divergence from control siRNAs, as identified by the ROUT method, are indicated by a green cross (x). Twelve genes known to be involved in secretion or protein trafficking were included in the screen as positive controls, indicated by a blue plus (+); these known controls were also identified as hits, validating the statistical method used here. ( B, C) Mean values for normalised luminescence for cell lysate and supernatant (SN) signals of the 92 hits identified. Here these hits are shown on a white-red scale, where darker red indicates a greater secretion defect. The generic GO_Slim was used to categorise hits as either related to protein transport, unrelated to protein transport or not annotated. ( B) A gene was defined as related to protein transport if it was annotated with any of the GO_Slim annotations transport (GO:0006810), transmembrane transport (GO:0055085) or vesicle-mediated transport (GO:0016192). ( C) Genes unrelated to protein transport or not annotated by GO_slim terms represent novel hits involved in protein secretion. Of the genes with no GO_Slim annotations, genes with no GO annotations at all are indicated in italics. Genes defined as not related to protein transport have other annotations in the generic GO_Slim.