Abstract
The original formulation of the string method in collective variable space is compared with a recent variant called string method with swarms-of-trajectories. The assumptions made in the original method are revisited and the significance of the minimum free energy path (MFEP) is discussed in the context of reactive events. These assumptions are compared to those made in the string method with swarms-of-trajectories, and shown to be equivalent in a certain regime: in particular an expression for the path identified by the swarms-of-trajectories method is given and shown to be closely related to the MFEP. Finally, the algorithmic aspects of both methods are compared.
Graphical abstract
1. INTRODUCTION
Understanding the mechanism by which activated processes such as conformational transitions and chemical reactions occurs in molecular systems is an issue of paramount importance in computational chemistry. Ideally, one would like to perform such analysis with a minimum of a priori assumptions,1,2 but this is often too complicated an aim. For example, brute-force molecular dynamics (MD) simulations are typically too slow to accrue enough statistics about these events, and the analysis of the output of such simulations can also prove quite complicated.3–5 In many situations, it is more practical to use a (possibly large) set of predefined collective variables (CVs) such as bond distances, dihedral angles, local density fields, etc. and calculate the transition paths in the CV space that the reaction is more likely to follow. In appropriate regimes, and when the right CVs are used, this strategy can indeed be justified and it can be shown that these transition paths are minimum free energy paths (MFEPs). The string method in collective variables6 is a simple and efficient technique to calculate MFEPs even in situations where the number of collective variables is very large (e.g., up to several thousands). The method represents the curvilinear reaction pathway between two free energy minima as a curve (the string) in the space of the collective variables. MD simulations of replicas of the full system restrained at the values of the collective variables at discretization points (or images) along the string are used to collect information about the system, and evolve the positions of these images until convergence. In its original formulation, one MD replica per image is used and the information needed is the mean force (gradient of free energy) and the value of a metric tensor, both of which can be expressed in terms of conditional averages. The string method has been employed in a variety of examples from MD.7–14 Recently,15 another version of the string method, termed string method with swarms-of-trajectories, has been proposed. Here, the idea is to update the positions of the images along the string using the average displacement in collective variables space of ensembles of short, free trajectories released from the current location of the images. The method has been used in refs16–18.
The main purpose of the present paper is to clarify the theoretical relationship between the original string method and the one using swarms of trajectories and to compare their practical implementation. As a main result, we give an expression for the path found by the swarms-of-trajectories method under specific conditions on the length of the underlying unbiased trajectories. We also revisit the assumptions underlying the string method and thereby discuss the significance of the MFEP in the context of reactive events.
2. THEORETICAL BACKGROUND
We will focus on a system of n particles evolving according to the Langevin equation,
| (1) |
where {(xi, vi)}i=1,…3n = (x, v) denotes the positions and velocities of the particles, mi is the atomic mass, U(x) the atomistic potential, γi the friction, kB the Boltzmann constant, T the temperature, and ηi(t) a white-noise term such that ⟨ηi(t)⟩ = 0 and ⟨ηi(t)ηj(t′)⟩ = δ(t − t′)δij. Other ensembles and/or thermal baths could be considered as well, but we will use the canonical ensemble and the Langevin dynamics for concreteness.
2.1. The Minimum Free Energy Path: Definition and Relevance
We start by recalling the definition of the MFEP then discuss its significance. Consider a set of collective variables (CVs), , that is, a vector-valued function that maps every configuration x of the system on a set of values (e.g., the dihedral angles along the molecule backbone or the distance between some of its atoms), and denote by the free energy (or potential of mean force, PMF) associated with these variables,
| (2) |
where is a normalization factor. By definition the MFEP is a curve in the space of the collective variables that connects the two minima A and B of and to which the vector is everywhere tangent;6 that is, it is a curve satisfying
| (3) |
where the superscript ⊥ indicates projection in the directions perpendicular to the curve. Here, is (minus) the mean force, that is, the gradient of the free energy, and M(z) is a metric tensor that accounts for the curvilinear nature of the CVs and guarantees invariance of the location of the MFEP under nonlinear transformations of these variables. The components of M(z) are given by the conditional average
| (4) |
or explicitly
| (5) |
As can be seen kBT M(z) has the dimension of a diffusion coefficient in CV space, that is, [kBT M(z)] = [z]2/[t].
Let us now explain why the MFEP is relevant to understand the mechanism of the transitions between a reactant state A and the product state B, where A and B are both appropriate subsets of ℝ3n × ℝ3n. For further details, we refer the interested reader to the original ref 6 and Appendix A. As shown in ref 19, in order to describe the statistical properties of the reactive trajectories by which reactive transitions occur, a key object is the committor function q(x,v). The committor function gives the probability that the trajectory initiated at (x,v) will reach first the product B rather than the reactant A, and it is the optimal reaction coordinate in that it enters the expressions for the probability density of reactive trajectories, their probability current, and the rate of the reaction. The committor function can be shown to satisfy a closed Fokker–Planck equation (see eq (A.1) in Appendix A). Unfortunately, this equation is too complicated to be solved, even numerically, by standard methods when the dimensionality of the system is large. To get around this difficulty, the main assumption made in the string method is that the committor function q(x,v) can be approximated by a function that does not depend on the velocities and only depends on the positions through the set of collective variables , that is,
| (6) |
As shown in ref 6 and recalled in Appendix A, by starting from a least-squares principle for q(x,v), we can then derive a simpler Fokker–Planck equation for Q(z) and observe that this is the equation for the committor function associated to the following overdamped equation for the collective variables
| (7) |
where γz is an artificial friction coefficient whose value is left unspecified by the argument. Equation 7 is central to the derivation of the MFEP, but its interpretation is quite subtle. Indeed, while this equation specifies some evolution rule for z(t), let us stress that we arrived at this equation without making any direct assumption about the dynamics of the CVs. Rather, the starting point is the ansatz (eq 6) for the committor function, which amounts to making a different assumption than those usually required to close the equations of motion at the level of the variable z (for example by assuming that these variables are slow compared to the rest). In particular, one can perfectly imagine situations in which the variables z are not slow, so that eq 7 does not capture the dynamics of these variables properly (this equation should rather be replaced by a non-Markovian generalized Langevin equation that could, for example, be derived via Mori–Zwanzig projection formalism20,21), and yet, the ansatz (eq 6) is accurate. What is the meaning of eq 7 in these situations then? Simply that the mechanism of the reaction can be understood through analysis of the solution of this equation; that is, that the reactive trajectories of the original system eq 1, once projected in collective variable space, follow the same channels as the reactive trajectories of eq 7. This is also why we can leave γz unspecified in eq 7, since its value does not change the location of the reaction channels. The philosophy here is somewhat analogous to that used in enhanced sampling methods for the calculation of the free energy associated with the collective variables: this free energy can be estimated by using a variety of dynamics, which are not necessarily identical to the original one in eq 1. Here, we are making a similar statement at the level of the channel of reactions viewed through the lens of the collective variables: these channels can be sampled using dynamics different from the original one in eq 1 (or its non-Markovian projection in CV space), and eq 7 is one of those.
Summarizing, the argument above implies that we can use eq 7 to study the mechanism of the reaction. This turns out to be relatively simple if we assume that the temperature is much smaller than the free energy barriers in the landscape that need to be crossed for the reaction to occur. Indeed, in these situations the reactive trajectories will concentrate with high likelihood in a thin tube centered around the MFEP. It is easy to understand why: if the temperature is low compared to the free energy barriers, we know that the transition state(s) will be the saddle point(s) on . On the way down from these saddle points, the thermal noise is not needed, and so the trajectory will simply follow the deterministic path solution of the zero-temperature version of eq 7
| (8) |
from the saddle point to the minimum of lying at the center of the product state. And since the dynamics in eq 8 is time-reversible (statistically), if we know the way down from the saddle point, we also know the way up to it: it will occur via the solution going from the minimum of lying at the center of the reactant state to the saddle state via the time-reversed of eq 8:
| (9) |
Taken together, the solutions to eqs 8 and 9 lie along a curve in collective variable space which connects the two minima of the free energy surface (A and B) via a saddle point and is everywhere tangent to the vector field ; that is, they lie along the MFEP. In essence, the string method is a way to identify the solutions to eqs 8 and 9 by thinking about them globally or geometrically and moving a curve in collective variable space using the location of the minima as only input. The algorithmic details are given in section 3.
Note that in both eqs 8 and 9 we could include the extra term in the drift involving kBT ∂Mα,β/∂zβ. In this case, the relevant path becomes the curve satisfying (compare eq 3)
| (10) |
where ∇·M(z) denotes the divergence of M(z), that is, the vector with components Σβ∂Mαβ/∂zβ. Because the extra term in 10 is proportional to kBT, it should not have much influence in the regime considered and the differences between the MFEP solution of eq 3 and the curve solution of eq 10 are expected to be small. This was indeed the result observed in ref 22 for the specific case of solvated alanine dipeptide, with dihedral angles as collective variables. Note, however, that the solution of eq 10 does not go exactly through the same critical points as the MFEP; that is, unlike the MFEP, it cannot be used to identify the saddle points on the free energy landscape.
The curve satisfying eq 10 is the one calculated by the on-the-fly string method,22 when this method is implemented using one system replica per string image (see eq 28 in that reference and Appendix B for more details). As we demonstrate next, eq 10 is also the path identified by the string method with swarms-of-trajectories, in the limit of short unbiased trajectories. It is also worth pointing out that the path defined by eq 10 is the one discussed recently in ref 23.
2.2. MFEP and the Swarms-of-Trajectories Method
In the original formulation of the string method in collective variables, the string, or parametrized curve representing the transition pathway, was evolved as a collection of images by estimating the mean force and the metric tensor at each image with constrained simulations. In contrast, the string method with swarm of trajectories uses eq 1 more directly to probe the reaction channel from the mean drift of the collective variables estimated on-the-fly via a large number (swarms) of short unbiased trajectories started at different points along the path. The main practical advantage is to bypass the need to explicitly calculate the mean force and the metric tensor from the conditional averages required in eqs 8 and 9. The idea was originally formulated by assuming that the collective variables z evolve according to noninertial Brownian dynamics on the free energy surface . In the following, we will present a geometric argument that is closer to the one made in the original string method paper.
The swarm method, as the original formulation of the string method, relies on the assumption that the reactive trajectories, once projected in collective variable space, go through a thin tube. Thus, if we were to initiate a trajectory anywhere in the thin tube of the reaction channel, then it should stay inside subsequently. In other words, if we pick a set of initial conditions (x(0), v(0)) such that is in the tube, then should also remain in the tube. Since all we observe are the CVs, we can as well consider the average over all initial conditions such that , that is, require that
| (11) |
remains in the tube if z was: here denotes the conditional average already used in eq 4. Since every z in the tube should lead through eq 11 to another value of the CVs, which is also in the tube, this tube should lie around a curve whose tangent is everywhere parallel to the vector parallel to the mean drift,
| (12) |
where Δt should be such is only a short drift distance away from z along the curve such that the vector can be used to obtain an accurate estimate of the tangent vector along the curve at position z. Successive iterations of the algorithm converge toward a curve whose tangent vector satisfies
| (13) |
as defined through the mean drift vectors via eq 12.
This equation calls for a few obvious questions. The first is what is the object that is computed this way and how does it compare to the MFEP solution of eq 3 or the curve solution of eq 10? As we show in the reminder of this section, the solution to eq 13 is in fact identical to that of eq 10 in the limit Δt → 0. The second question is how do the two string methods, the original one and the one with swarms, compare from an algorithmic viewpoint: this question will be addressed in section 3.
Let us now prove that the solution to eq 13 is identical to that of eq 10 in the limit Δt → 0. Suppose that the propagation time Δt of the unbiased trajectories in the swarms is very short. In this case, we have
| (14) |
where vi denotes the velocity of the Cartesian variables. We can then estimate for small Δt by computing the canonical average of , with given by eq 14. Since the average of the first order terms in eq 14, , is zero, we arrive at
| (15) |
where Aα(z) is given by the conditional average,
| (16) |
(note that the atomic friction coefficients, γi, have disappeared from the expression). Equation 16 can be rewritten as
| (17) |
or more explicitly,
| (18) |
with and Mαβ(z) given by eqs 2 and 4 respectively. This calculation shows that the vector with components Aα(z), and hence the vector in the limit as Δt → 0, is parallel to the vector , thereby justifying that the curve solution of 13 in the limit Δt → 0 is the same as the curve solution of eq 10.
The argument above does not say what is for larger value of Δt. From the discussion that led to eq 13; however, we can infer that using larger values of Δt (but still small enough that and z remains close along the curve) will have the effect of smoothing the curve.
3. ALGORITHMIC ASPECTS
In this section, we recall the operational details of the mean forces string method and then discuss the swarms-of-trajectories approach. A summary of the practical steps performed in the two algorithms is reported in Table 1.
Table 1.
Operational Comparison of the Mean Forces and Swarms-of-Trajectories String Methodsa
| mean forces | swarms of trajectories | |
|---|---|---|
| 0 | build initial path | build initial path |
| 1 | restrained sampling to estimate mean forces | short restrained sampling to prepare swarms |
| 2 | run swarms | |
| 3 | evolve collective variables with estimate of mean forces | evolve collective variables with estimate of drift |
| 4 | reparametrization | reparametrization |
Steps 1 to 4 are performed at each image and iterated until convergence.
3.1. String Method with Mean Forces
The string method in collective variables space is a generalization of the string method in Cartesian space, an algorithm designed to find Minimum Energy Paths (MEPs) by evolving a curve (the string) while maintaining its parametrization. The essence of the method is to discretize the string into M + 1 points, zi, with i = 0,1,…,M, which are evolved by iterating over the following two steps until convergence:
1. Evolution
All images are evolved independently from each other using the following updating rule:
| (19) |
where h > 0 is a parameter with the dimension of a time divided by a friction coefficient (i.e., a time square), the choice of which is discussed below along with the way to calculate .
2. Interpolation/reparametrization
A piecewise linear curve is interpolated though the images and new images are distributed along this curve in such a way that they are equidistant from each other. To this end, we first calculate the piecewise linear function l(s), s ∈ [0, 1], such that l(0) = 0 and
| (20) |
where |·| denotes Euclidean norm. Then, we pick new images along the curve at the new parameter values si specified by
| (21) |
This guarantees that these new images satisfy
| (22) |
to leading order in 1/M.
At convergence, the images slide along the curve due to the evolution step, then go back to their positions at the beginning of the loop due to the interpolation/reparametrization. This means that the algorithm identifies a curve that is left invariant by the flow , that is, a MFEP defined by eq 3.
We now describe how to compute and to choose h. Since M(zi) and are defined via conditional expectations at (see for example the expression for M(z) in eq 5), they can be estimated by using holonomic constraints and the Blue Moon ensemble method.24 However, it is often simpler in practice to approximate the constraint with a stiff restraining potential. To each image zi, we associate an independent full-atom replica of the system, xi, whose evolution we simulate by adding to the standard MD force-field the restraint potential
| (23) |
Using restraints versus constraints implies that the motion of the collective variables occurs on a smoothed version of the true free energy, , defined as (compare eq 2)
| (24) |
where is a normalization factor. Indeed, it can be seen that
| (25) |
where is the free energy defined in eq 2. Hence, kBT/κ is the variance of the smoothing filter and its square root should be smaller than the scale at which we want to resolve .
Denoting by xi(t) the MD trajectory of the replica assigned to image zi, we use the following estimators for the αth component of the free energy gradient and the (α,β)th entry of the tensor M(zi)
| (26) |
| (27) |
where T is a time interval. The quality of the approximations in eqs 26 and 27 depends on κ and T and has been analyzed in ref 6.
Equations 19, 26, and 27 suggest that h must be chosen small enough so that the evolution step in eq 19 is stable and accurate, and T large enough so that the time averages in eqs 26 and 27 are converged. If such values of h and T have been identified, however, it was shown in ref 22 that we can then decrease T to a value as low as that of the time-step δt in the MD calculation (for example δt ≈ 1 fs) provided that we scale h accordingly by taking h ∝ δt/T. In this regime, the images zi evolve much more slowly than the corresponding MD replicas and effectively feel the average effect of the MD replica. The advantage is that this makes the evolution of the images zi and the replica xi concurrent, and hence, no reinitialization of the MD is required. This idea is at the basis of the on-the-fly string method which is summarized in Appendix B.
Finally, notice that a lower force constant κ in eq 23 can also be used to accelerate sampling for example by combining with replica exchange techniques (see for example ref 9).
3.2. String Method with Swarms-of-Trajectories
As we have discussed in section 2.2, the swarms-of-trajectories version of the string method aims at identifying a path defined by eq 13 rather than eq 3. In practice, this can be done by an algorithm similar to the one described in section 3.1 in which the evolution step is replaced by the following:
1′. Evolution
All images are evolved independently from each other using the following updating rule:
| (28) |
where r is a dimensionless rescaling factor which in practice can be taken to be 1.
To estimate , we proceed as follows. To each image zi we associate an ensemble of S initial conditions such that , k = 1,…,S. This ensemble of initial conditions can be generated for example using MD simulations in a restraining potential such as the one in eq 23. These initial conditions are then let free to evolve by unrestrained MD for a time Δt, and we estimate the average displacement for the collective variables as
| (29) |
The approximation in eq 29 is controlled by the number of trajectories S we use in the swarms, while the length of the time interval Δt affects the path identified by the definition eq 13.
The update in eq 28 is performed on all the images along the string, thus providing an updated set of images that are successively made equidistant by applying the interpolation/reparametrization step to enforce eq 22. Finally, to prepare the S initial conditions for the next iteration one can use as starting conformation the one among the xi,k(Δt) such that is the closest to the updated position of the image zi. Notice, however, that unlike the mean force version of string method, the swarms-of-trajectories version always requires reinitialization.
4. ILLUSTRATIVE EXAMPLES
4.1. Mueller Potential
In this section, we use a simple numerical example to illustrate the features of the swarms-of-trajectories string method and compare it with the original string method. Specifically, we consider a 2-dimensional system evolving according to Langevin dynamics on the Mueller potential (see Figure 1) and take the coordinates of the system as collective variables, that is, – with this choice the MFEP reduces to a MEP on the potential. We set the friction coefficient to γ = 100, which is about twice the average value of the square root of the eigenvalues of the Hessian computed at the minimum of the potential. The Langevin equations of motion are integrated with the second order algorithm of ref 25 with time step δt = 10−4, mass 5, and kBT = 10.
Figure 1.
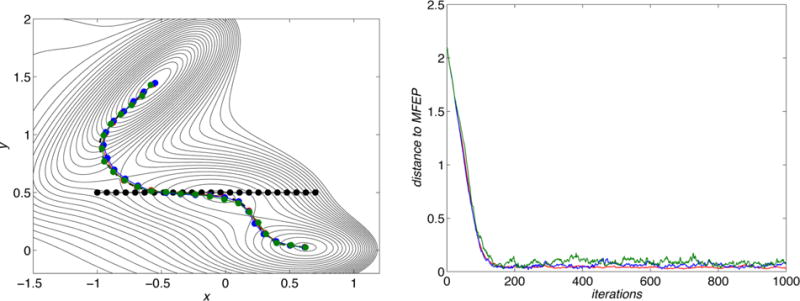
Convergence of the original string method. Left panel: Mueller potential surface with the initial string (black circles), the MFEP (black dashed line), and the final strings (circles) obtained using different values of the noise in the determination of the (mean) forces. The noise is represented by adding to each force a normally distributed random number of increasing standard deviation, blue = 10, red = 30, and green = 50. Right panel: distance from the MFEP during the evolution of the string with increasing noise.
We begin by showing the results of the original string method. As mentioned in section 3.1, in general the error in the estimation of mean forces has two contributions. One is systematic and depends on the choice of κ in eqs 23 and 26, while the other one is statistical and due to finite sampling effects, that is, the length of the MD trajectories. Explicit expressions of the different contributions can be found in the Appendix of ref 6. The statistical error can be made as small as desired by running longer trajectories, and the systematic error can be eliminated by using constrained MD as in ref 24 or reduced by choosing a larger κ. In the example described here, the mean force is the force and we will assume that there is no systematic errors in its calculation (as if κ was large enough). However, we mimic the statistical errors by adding to each force a random number normally distributed with mean 0 and different standard deviations of 10, 30, and 50 (shown respectively in blue, red, and green in Figure 1). These values are about 5%, 15%, and 25%, respectively, of the maximum value of the force at the string images during its evolution. In the left panel of Figure 1, we show the Mueller potential surface with the initial string represented as black circles, the MFEP as a black dashed line, and the converged strings obtained at the different values of the error. Strings of 24 images were considered. In the right panel of the figure, we show the distance from the MFEP during the iterations made to evolve the string: this gives an estimate of both the speed of convergence and the remaining error at convergence due to the statistical error. As can be seen, the error in the force calculation do not alter the convergence rate but they increase the fluctuations around the converged path. However, these fluctuations remain small for all the values of the statistical error that we considered.
Next, we analyze the results of the swarms-of-trajectories string method. In this case, the sources of errors are the number of trajectories in the swarms, the initialization (whether one uses restraints or constraints), and the length of the unbiased trajectories in the swarms. In our experiments, we observed that the error is dominated by the effect of the length of the unbiased trajectories, and hence, we only show here how the path determined by the algorithm varies when we change this parameter while keeping fixed the others. We use swarms of 103 trajectories and to mimic the initialization error we generate the initial coordinates for the swarms trajectories randomly, using a normal distribution with mean zi and standard deviation 0.005 in both dimensions. We use again 24 images to represent the strings and a scaling factor r = 1 for images evolution (see eq 28). The left panel of Figure 2 shows the final path obtained using different values of the time-lag Δt, that is, a different number of time-steps in the free trajectories (red = 100, blue = 1000, green = 5000, orange = 8000), while the right panel shows the distance from the MFEP versus the total number of steps during the string evolution (computed as the number of steps per swarm trajectory times the number of iterations).
Figure 2.
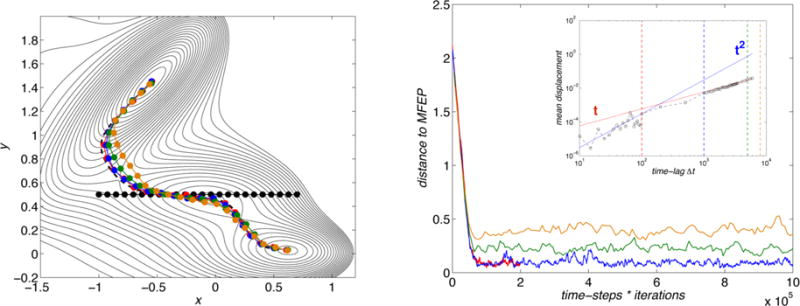
Convergence of the swarms-of-trajectories method. The left panel shows the Mueller surface with the initial string (black circles) and the MFEP (black dashed line), together with the final paths (circles) obtained using 103 trajectories in the swarms and different time lags Δt (red = 100 steps, blue = 1000, green = 5000, orange = 8000). Right panel: distances from the MFEP as a function of the total number of steps during the evolution of the strings, using the same color coding as in the left panel. The inset shows how the mean displacement of the trajectories in a swarm varies when increasing the time lag Δt: the red and blue solid lines indicate linear and quadratic behaviors, respectively, while the vertical dashed lines show the time lag Δt used with the same color code of the main panels. In this calculation, the trajectories were started from the point (x, y) = (−1.2, 1.4) on the Mueller surface.
The inset in the figure also shows the way the mean displacement of the trajectories in the swarm varies with the length of the free trajectories. When the displacement is quadratic in Δt, we are in the regime considered at the end of section 3.2 (see eq 15) where the curve identified by the swarms-of-trajectories string method is (close to) the MFEP. At longer lag, however, the displacement becomes linear in Δt, and the calculation at the end of section 3.2 is no longer valid. Indeed, it can be seen from the figure that, as the lag Δt increases, the curve identified by the swarms-of-trajectories string method becomes increasingly different from the MFEP. The reason behind this discrepancy can be understood from Figure 3, where we show the spread of the trajectories in the swarms by plotting the final positions (xi,k(Δt), yi,k(Δt)) of 103 trajectories launched from an image along the converged path (image number 10, which is close to the saddle point) for the various values of the lag time Δt. As the lag time increases the free trajectories used in the swarm spread more and more toward the closest minima, splitting among the two sides of the potential surface divided by the saddle point. This effect also explains the corner-cutting effect along the path that is observed in the left panel of Figure 2 for the larger values of Δt.
Figure 3.
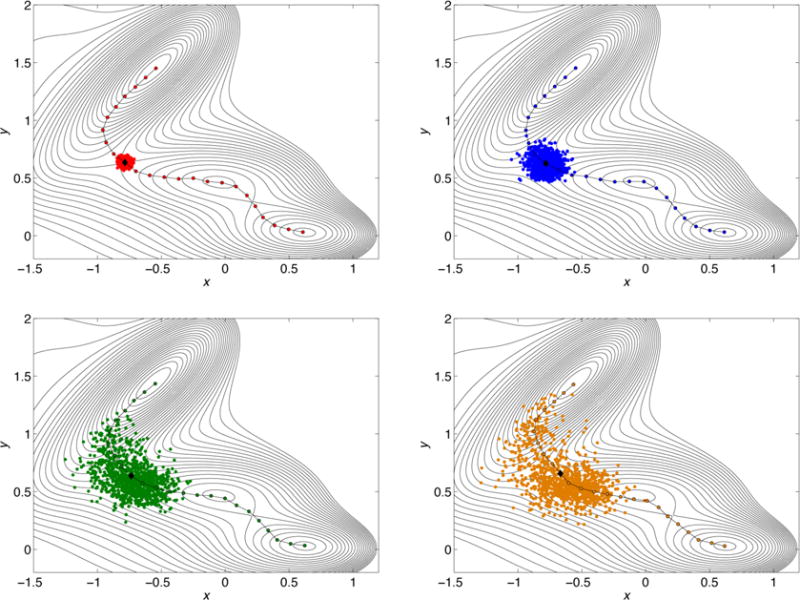
Spread of the free trajectories in the swarm around a selected image on the converged path at different trajectories length. Going from left to right and top to bottom it is shown the spread at different time lags Δt: the number of MD steps in the free swarm trajectories are 100, 1000, 5000, and 8000, respectively, and the color code is the same of Figure 2. The image chosen is indicated with a black diamond and it is an approximation of the saddle point in the different strings.
4.2. Alanine Dipeptide
In this section we analyze the performance of three different implementations of the string method on a prototypical biomolecular testcase, the isomerization transition of an alanine dipeptide molecule in vacuum. We study the performance of three different algorithms, the original one based on averaged mean forces, the swarms-of-trajectories and the on-the-fly method. The on-the-fly string method was first introduced in ref 22, and for completeness, it is summarized in Appendix B. As recalled in section 2, while the original mean forces string method provides the MFEP defined by eq 3, the swarms-of-trajectories method with short time-lag and the on-the-fly method with one replica per image both converge to the path defined in eq 10.
We employ the algorithms to investigate the transition between the dipeptide metastable conformers named C7eq and C7ax by using the two backbone dihedral angles as collective variables, i.e. (z1, z2) = (φ, ψ). Although it is known that the angles around the peptide bonds must also be included to properly describe the mechanism of the transition,9,26,27 here we use only the backbone angles for illustrational purposes. The values of these dihedrals at the metastable conformers are (φ, ψ)C7eq ≈ (−85,75) and (φ, ψ)C7ax ≈ (70, −70). All the scripts used for the calculations presented in this section are provided as Supporting Information. MD simulations were performed using the NAMD program28 and the CHARMM27 force field.29 For the alanine dipeptide molecule we used Langevin dynamics at 300K with a time step 1 fs and a friction coefficient of 10.0 ps−1 on all atoms. The simulations were run in vacuum with dielectric constant 1.
In all cases, as initial string we used a linear path connecting the points (−80, 75) and (50, −100) and discretized using 20 images. For the averaged mean forces method we estimated the forces and the metric tensor using eqs 26 and 27 and restrained trajectories of T = 20 000 fs and κ = 1000 kcal/mol·rad2, and a step h = 0.02 for the evolution of the images (see eq 19). For the swarms-of-trajectories version we used κ = 1000 kcal/mol·rad2 and 10 000 fs to prepare the initial condition for the swarms at each image, and then launched 100 free MD trajectories of 20 fs lag Δt each. Hence, the total cost per image and per iteration is 12 000 MD steps of 1 fs (i.e., 12 ps). A scaling factor r = 1 (see eq 28) was used for images evolution. We note that 20 fs of free trajectories for the swarms means that we are in the short time-lag regime. Since for the present application we have implemented all the algorithms serially, it becomes costly to run longer trajectories. For more complex molecular examples to be run on supercomputers, however, it is convenient to submit all the calculations needed to update each image in parallel. In this setup, longer unbiased trajectories can be used. It is not clear if this is useful, however, since we know from the previous section that using longer trajectories modifies the path. Finally, for the on-the-fly method we used again κ = 1000 kcal/mol·rad2 and a ratio for the friction coefficients γz/γi = 100. The images were reparametrized each 500 fs.
Figure 4 shows the converged paths from the different algorithms, plotted on the free energy landscape for the (φ, ψ) angles calculated using the single-sweep method30 (the zero of the free energy is at (φ, ψ) = (−80, 70) and the levels are drawn at 1.1 kcal/mol). We show in red the MFEP defined by eq 3 and obtained using the original string method with averaged mean forces, in green the path obtained using the swarms-of-trajectories method and in blue the path from the on-the-fly method. The swarms-of-trajectories method with short time-lag and the on-the-fly method both converge to the path described by eq 10. It can be observed that in the present application this path differs only slightly from the MFEP, as it was already recognized for the alanine dipeptide in solution.22 Figure 5 shows the computational cost for the three different string method algorithms, calculated as the distance from the initial path versus the total number of per-image MD steps performed during the evolution of the string. The color code is the same as that of Figure 4. We note that the on-the-fly method provides the same path of the swarms-of-trajectories in a smaller number of steps, but the latter does not require the explicit calculation of the tensor in eq B.2.
Figure 4.
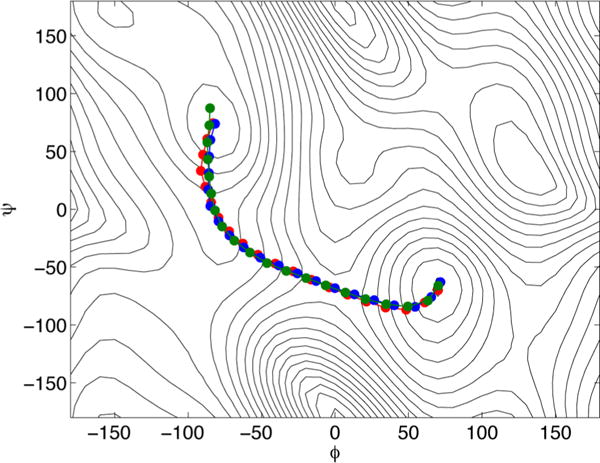
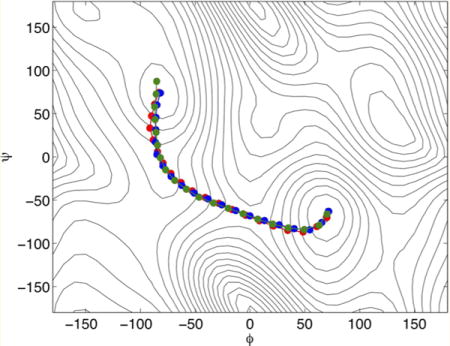
Converged paths for the alanine dipeptide test obtained using the different string method algorithms: the original method using averaged mean forces (the MFEP, red), the swarms-of-trajectories (green), and the on-the-fly method (blue).
Figure 5.
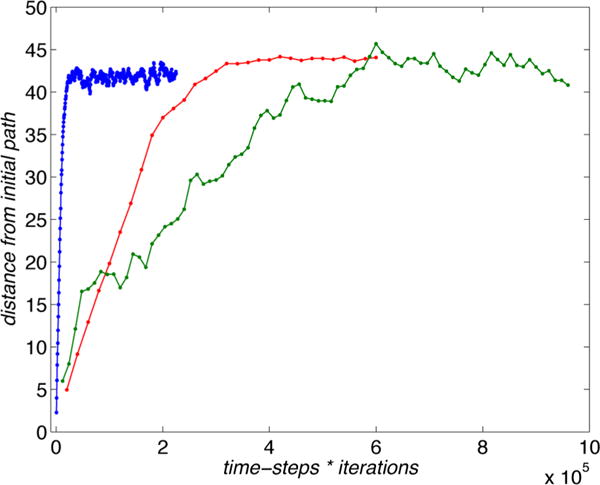
Computational cost of the different string method algorithms for the alanine dipeptide test: distance from the initial path as a function of the total number of steps during the evolution of the string. The original method using averaged mean forces (red), the swarms-of-trajectories (green), and the on-the-fly method (blue).
5. CONCLUDING REMARKS
We have compared the original formulation of the string method in collective variable space with a more recent version using swarms of trajectories. It was shown that this second method computes a path generally close to the MFEP, at least if the lag time in the swarm is chosen short enough. The practical implementation of the methods were also compared and illustrated on a simple two-dimensional problem and the MD example of the alanine dipeptide molecule.
One open question that remains is what path does the string method with swarms-of-trajectories compute if longer lags are used. The longer the lag, the straighter the path, which may be useful in practice, especially if the free energy landscape is somewhat rugged. Indeed, using longer lags in effect amounts to smoothing the small scale features on this landscape. How to precisely quantify this smoothing effect and connect it, for example, to the thermal averaging performed in the finite-temperature string method31,32 or to an homogenization step where the true propagation of the microscopic system is replaced by a simpler approximate system with renormalized drift and diffusion coefficient33 remains to be done. Such an analysis may provide a better understanding of the quality of the ansatz in eq 6, and perhaps lead to a more accurate redefinition of the tensor M(z) in eq 4.
Acknowledgments
We thank Albert C. Pan and Deniz Sezer for stimulating discussions. The research of B.R. was funded by grant MCB-0920261 from the National Science Foundation (NSF). The research of E.V.-E. was supported in part by National Science Foundation grant DMS07-08140 and Office of Naval Research (ONR) grant N00014-11-1-0345.
APPENDIX
A. Derivation of eq 7
As shown in ref 19 and already mentioned in section 2.1, the key object needed to describe the statistical properties of the reactive trajectories by which such transitions occur is the committor function q(x,v). The committor function is known to satisfy the following Fokker–Planck equation:
| (A.1) |
to be solved with the boundary conditions that q = 0 on ∂A and q = 1 on ∂B. Unfortunately, eq A.1 is a very complicated equation that cannot be solved, even numerically, by standard methods when the dimensionality of the system is large. Assume now that the committor function q(x, v) can be approximated by a function that only depends on the positions through the set of collective variables , that is, assume that eq 6 holds. The question then becomes how to reduce A.1 into an equation for Q(z), and how to analyze this equation. In the original string method in collective variables paper,6 this is done via a least square principle. Clearly, the solution to eq A.1 minimizes the following objective function
| (A.2) |
where H is the Hamiltonian
| (A.3) |
If we substitute the ansatz into eq A.2, it is easy to see after a few manipulations that it reduces to the following objective function for Q(z):
| (A.4) |
where is the free energy defined in eq 2 and the tensor Mα,β(z) is given by the conditional average eq 4. The Euler–Lagrange equation associated with the minimization of eq A.4 is another Fokker–Planck equation:
| (A.5) |
If N is large (more than 3, for example), this equation is again difficult to solve, but it permits to reformulate the problem in a convenient way. Indeed, A.5 is precisely the equation for the committor Q(z) associated with the overdamped equation for the variables z given in 7. Note also that this explains why the artificial friction γz is left unspecified in eq 7, since the physical time does not enter A.5 any longer.
B. On-the-Fly String Method
In the on-the-fly string method with one replica per image we evolve the images along the string concurrently with a set of replicas of the MD system, without ever needing to reinitialize the system nor compute mean forces. It is useful to consider the continuous version of the method, in which case the evolving string is a time-dependent curve parametrized as {z(s,t): s ∈ [0,1]} which one evolves concurrently with a one-parameter family of replicas of the MD system, {x(s,t): s ∈ [0,1]}. Written component-wise for α = 1,…,N and i = 1,…,3n, the equations for the concurrent dynamics are
| (B.1) |
where γz is an effective friction term acting on the collective variables and
| (B.2) |
In B.1, the term , with , is used to enforce the particular parametrization chosen for the curve (for example by normalized arclength).
In ref 22, it was shown that if we simulate B.1 with κ and γz sufficiently large the equation for zα(s,t) in B.1 can be replaced by the following effective equation
| (B.3) |
As can be seen, the steady state solution of B.3 is exactly 10. This means that if we evolve a string {z(s): s ∈ [0, 1[} by simulating B.1 using large but finite values of κ and γz, this will converge to the curve defined by eq 10.
Footnotes
Supporting Information
Scripts used for the calculations presented in section 4.2. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
References
- 1.Bolhuis PG, Chandler D, Dellago C, Geissler PL. Annu Rev Phys Chem. 2002;53:291–318. doi: 10.1146/annurev.physchem.53.082301.113146. [DOI] [PubMed] [Google Scholar]
- 2.Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 3.Tu T, Rendleman CA, Borhani DW, Dror RO, Gullingsrud J, Morten, Klepeis JL, Maragakis P, Miller P, Stafford KA, Shaw DE. SC ’08: Proceedings of the 2008 ACM/IEEE Conference on Supercomputing. 2008:1–12. [Google Scholar]
- 4.Krivov SV. J Phys Chem B. 2011;115:12315–12324. doi: 10.1021/jp208585r. [DOI] [PubMed] [Google Scholar]
- 5.Best RB, Hummer G, Eaton WA. Proc Natl Acad Sci USA. 2013;110:17874–17879. doi: 10.1073/pnas.1311599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maragliano L, Fischer A, Vanden-Eijnden E, Ciccotti G. J Chem Phys. 2006;125:024106. doi: 10.1063/1.2212942. [DOI] [PubMed] [Google Scholar]
- 7.Miller TF, Vanden-Eijnden E, Chandler D. Proc Natl Acad Sci USA. 2007;104:14559–14564. doi: 10.1073/pnas.0705830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu F, Hummer G. Proc Natl Acad Sci USA. 2010;107:19814–19819. doi: 10.1073/pnas.1009313107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ovchinnikov V, Karplus M, Vanden-Eijnden E. J Chem Phys. 2011;134:085103. doi: 10.1063/1.3544209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirmizialtin S, Nguyen V, Johnson KA, Elber R. Structure. 2012;20:618–627. doi: 10.1016/j.str.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stober ST, Abrams CF. J Phys Chem B. 2012;116:9371–9375. doi: 10.1021/jp304805v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vashisth H, Maragliano L, Abrams CF. Biophys J. 2012;102:1979–1987. doi: 10.1016/j.bpj.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsunaga Y, Fujisaki H, Terada T, Furuta T, Moritsugu K, Kidera A. PLoS Comp Biol. 2012;8:e1002555. doi: 10.1371/journal.pcbi.1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vashisth H, Abrams CF. Proteins: Struct Funct Bioinf. 2013;81:1017–1030. doi: 10.1002/prot.24255. [DOI] [PubMed] [Google Scholar]
- 15.Pan AC, Sezer D, Roux B. J Phys Chem B. 2008;112:3432–3440. doi: 10.1021/jp0777059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gan W, Yang S, Roux B. Biophys J. 2009;97:L8–L10. doi: 10.1016/j.bpj.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jo S, Rui H, Lim JB, Klauda JB, Im W. J Phys Chem B. 2010;114:13342–13348. doi: 10.1021/jp108166k. [DOI] [PubMed] [Google Scholar]
- 18.Lacroix JJ, Pless SA, Maragliano L, Campos FV, Galpin JD, Ahern CA, Roux B, Bezanilla F. J Gen Phys. 2012;140:635–652. doi: 10.1085/jgp.201210827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanden-Eijnden E. In: Computer Simulations in Condensed Matter Systems: From Materials to Chemical Biology Vol 1. Ferrario M, Ciccotti G, Binder K, editors. Springer; Berlin: 2007. pp. 453–493. [Google Scholar]
- 20.Grabert H. Projection Operator Techniques in Nonequilibrium Statistical Mechanics. Springer Verlag; Berlin: 1982. pp. 9–26. [Google Scholar]
- 21.Chorin AJ, Hald OH. Stochastic Tools in Mathematics and Science. Springer; New York: 2005. pp. 145–149. [Google Scholar]
- 22.Maragliano L, Vanden-Eijnden E. Chem Phys Lett. 2007;446:182–190. [Google Scholar]
- 23.Johnson ME, Hummer G. J Phys Chem B. 2012;116:8573–8583. doi: 10.1021/jp212611k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciccotti G, Kapral R, Vanden-Eijnden E. ChemPhysChem. 2005;6:1809–1814. doi: 10.1002/cphc.200400669. [DOI] [PubMed] [Google Scholar]
- 25.Vanden-Eijnden E, Ciccotti G. Chem Phys Lett. 2006;429:310–316. [Google Scholar]
- 26.Bolhuis PG, Dellago C, Chandler D. Proc Natl Acad Sci USA. 2000;97:5877–5882. doi: 10.1073/pnas.100127697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma A, Dinner AR. J Phys Chem B. 2005;109:6769–6779. doi: 10.1021/jp045546c. [DOI] [PubMed] [Google Scholar]
- 28.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-Mccarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 30.Maragliano L, Vanden-Eijnden E. J Chem Phys. 2008;128:184110. doi: 10.1063/1.2907241. [DOI] [PubMed] [Google Scholar]
- 31.E W, Ren W, Vanden-Eijnden E. J Phys Chem B. 2005;109:6688–6693. doi: 10.1021/jp0455430. [DOI] [PubMed] [Google Scholar]
- 32.Vanden-Eijnden E, Venturoli M. J Chem Phys. 2009;130:194103. doi: 10.1063/1.3130083. [DOI] [PubMed] [Google Scholar]
- 33.Pavliotis G, Stuart A. Multiscale Methods: Averaging and Homogenization. Springer; New York: 2008. pp. 163–186. [Google Scholar]


