Fig. 7. Model for substrate processing by the Cdc48 ATPase complex.
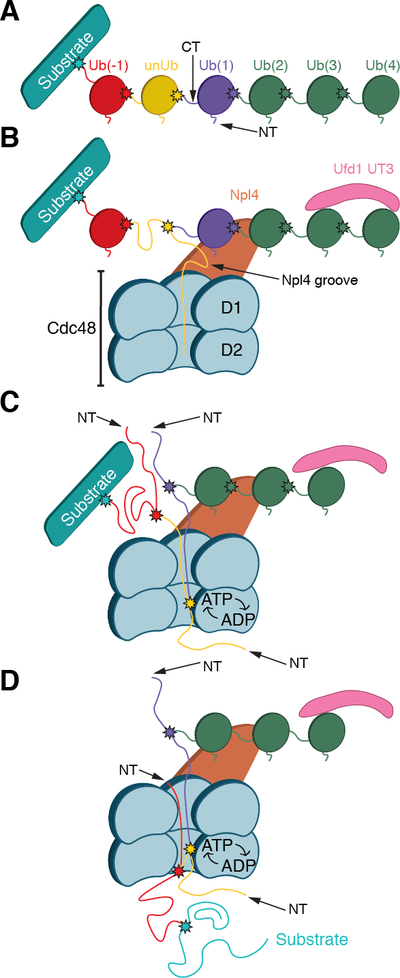
(A) Scheme of a substrate with an attached Lys48-linked poly-ubiquitin chain. In the example chosen, the ubiquitin molecule shown in yellow will be unfolded; it is separated from the substrate by one ubiquitin molecule (Ub(−1)). Ubiquitins distal to the one being unfolded are numbered (Ub(1)…Ub(4)). Lys48 branch points are indicated by stars. (B) Binding of the substrate to the Cdc48 complex. The distal folded ubiquitins Ub(1) and Ub(2) bind to the top of Npl4, and Ub(3) and possibly Ub(4) bind to the UT3 domain of Ufd1. The unfolded ubiquitin (unUb) binds to the groove in Npl4 and projects its N-terminus across both ATPase rings. (C) ATP hydrolysis in the D2 ring pulls the N-terminus of the unfolded ubiquitin through the central pore, moving the branch point into the ATPase rings and causing unfolding of attached Ub(1). Pulling on the proximal side of the ubiquitin chain results in unfolding of Ub(−1). (D) Ultimately the substrate is moved through the central pore and unfolded. Substrate release from the ATPase complex requires the removal of the distal ubiquitins Ub(2)-Ub(4) by a DUB.
