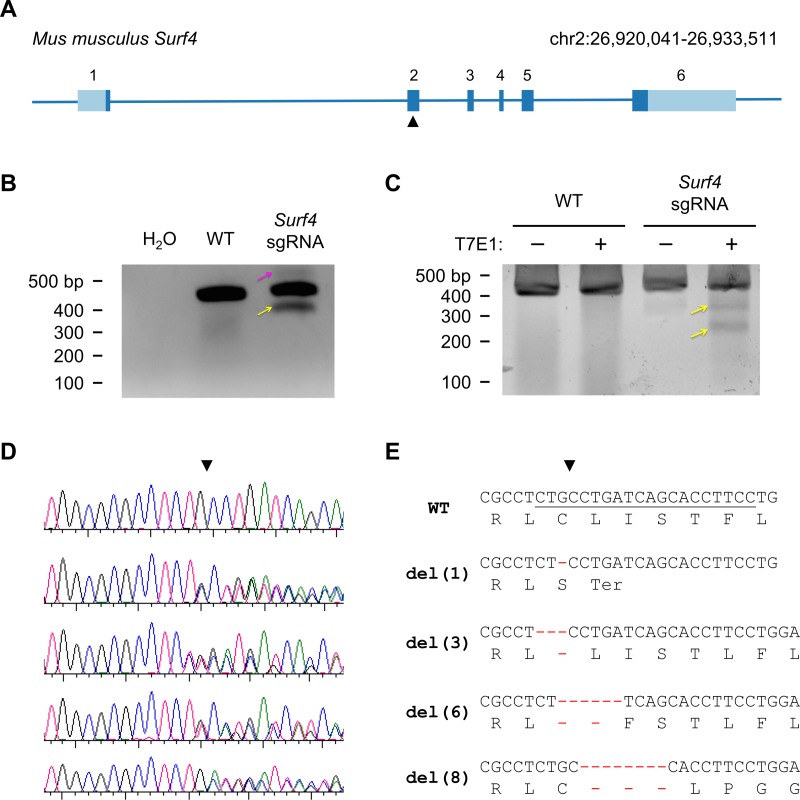
Fig 1. Generation of Surf4 mutant alleles.
(A) Surf4 gene structure. Exons are shaded light blue for untranslated regions or dark blue for coding sequence. The target site for the sgRNA used for oocyte editing is indicated by the black triangle. (B) Mouse ES cells were either untreated or electroporated with plasmids for CRISPR/Cas9 disruption of the Surf4 target site. PCR amplification of genomic DNA or water control across the Surf4 target site revealed higher and lower molecular weight DNA fragments suggestive of nonhomologous endjoining repair of Surf4 indels. (C) The major PCR product was gel purified and subjected to T7 endonuclease I digestion. T7E1 digestion produced novel DNA fragments (arrows) indicating the presence of insertions/deletions in Surf4 exon 2. Wild type DNA was resistant to T7E1 digestion. (D) Sanger sequencing chromatograms of Surf4 target site amplicons of progeny from matings between Surf4-targeted founder mice and wild-type C57BL6/J mice. (E) DNA and predicted protein sequences for the 4 individual allele generated by CRISPR/Cas9 gene-editing of Surf4.