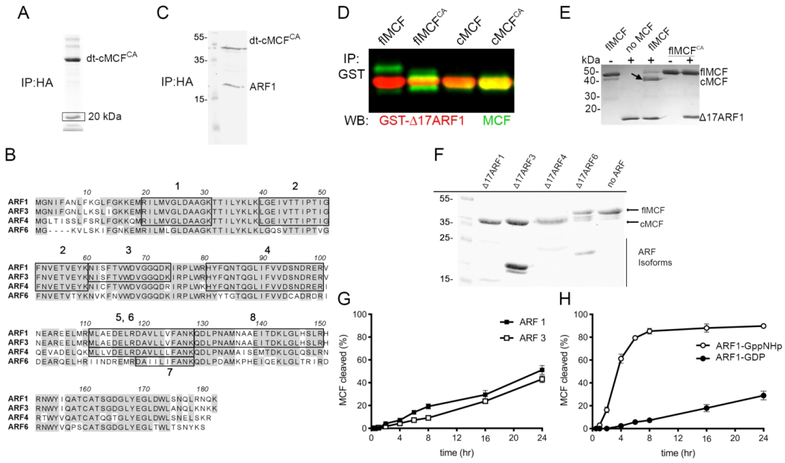
Figure 1.
MCF autoproteolytic cleavage is induced by ARF GTPases.
A. dt-cMCFCA was immunoprecipitated (IP) from HEK293T cell lysate using anti-HA antibody. Box indicates band excised for mass spectrometry peptide sequencing. B. Amino acid sequence alignment of major isoforms of ARFs with boxes indicating identified peptides. Peptides marked 6 and 7 are unique to ARF4 and ARF6, respectively. C. Immunoblot showing endogenous ARF1 pulls-down with dt-cMCFCA recovered from whole cell lysate as detected by anti-ARF1 antibody and anti-HA. Replicate pull-downs of independent transfections can be found in Supplemental Fig. 1. D. Purified GST-Δ17 ARF1 was incubated with either MCF, MCFCA, cMCF, or cMCFCA at 37°C. Western blot on samples following anti-GST IP using anti-MCF (green bands) and anti-ARF1 (red bands). showing catalytically active and inactive flMCF and cMCF (green bands) can bind ARF1(red bands) in vitro. E. Recombinant flMCF and flMCFCA incubated with ΔARF1 indicate induced autoprocessing is dependent on MCF catalytic active site. Arrow indicates cleaved MCF band. F. Auto-cleavage induced by purified ARF isoforms incubated with MCF for 24 hours. G, H. Autoprocessing of flMCF at 37°C for time indicated induced by full-length ARF1 or ARF3 (F) or with ARF1 pre-loaded with GDP or non-hydrolyzable GTP (GppNHp) (G). Representative gels (n>3), with percent cleaved MCF was determined by densitometry of bands on Coomassie stained gels (%=(cMCF/(cMCF + flMCF)*100)) from three independent reactions.