Fig. 1. The environmental and genetic determinants of plasmid mobility.
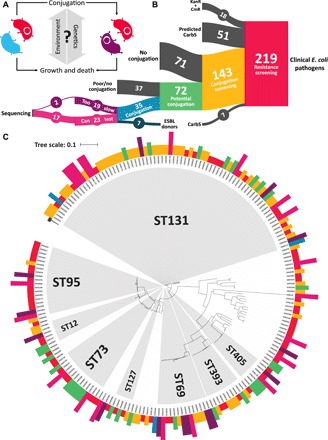
(A) The influence of environmental and genetic factors on conjugation is confounded by growth dynamics between donor (red), recipient (blue), and transconjugant (purple) populations. By decoupling conjugation modulation from growth dynamics, we can identify and use the determinants of plasmid mobility to fight antibiotic resistance. (B) Assembling a library of natural isolates with quantifiable rates of conjugation is a substantial undertaking. Starting from a library of 219 clinical E. coli pathogens from patient bloodstream infections, we screened for the ability to transfer β-lactam resistance commonly found on plasmids native to the Enterobacteriaceae family. Approximately 25% of the carbenicillin-resistant (CarbR) isolates exhibited detectable transfer to chromosomally kanamycin (KanR)– or chloramphenicol (CmR)–resistant recipients. These and seven extended spectrum β-lactamase (ESBL) donors were subsequently used to examine environmental and genetic determinants of plasmid mobility. (C) The diversity present in the E. coli pathogen library is maintained through conjugation screening. A phylogenetic tree of the library was constructed from 200 genome assemblies (BioProject accession nos. PRJNA290784 and PRJNA551684) to reveal the breadth of our analysis throughout each phase of screening. Genome assemblies for the remaining 19 isolates were either unavailable or of insufficient quality. E. coli strain EC958 (GenBank accession no. HG941718.1) was used as a reference genome for alignment. Isolates are color labeled by their final phase. Major multilocus sequence types (≥5 isolates in common) present in the library are highlighted in gray.
