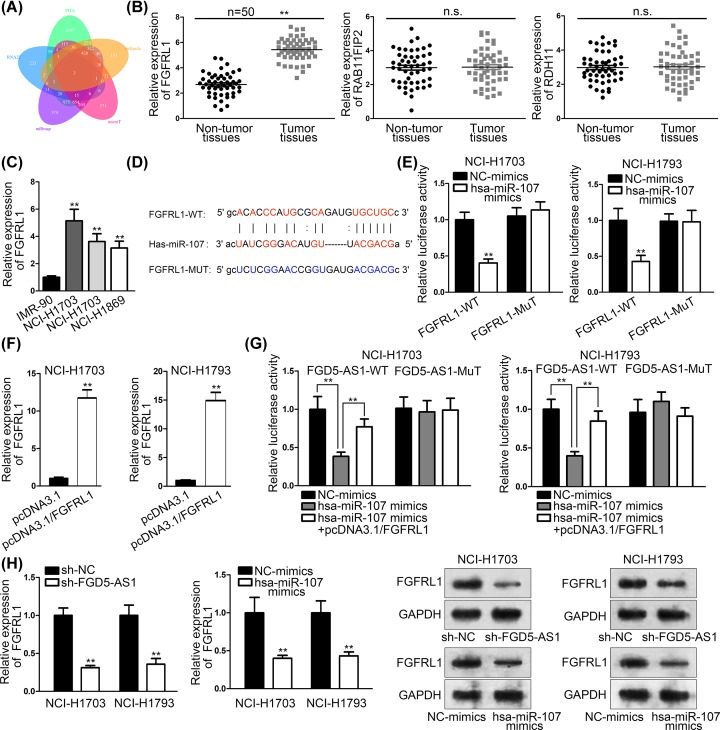
Figure 3. Hsa-miR-107 targeted FGFRL1.
(A) StarBase was used to predict potential mRNA. (B) The expressions of three different types of mRNA were detected by qRT-PCR assays. (C) qRT-PCR assays were used to detect the expressions of FGFRL1 in cell lines (IMR-90, NCI-H1703, NCI-H1793 and NCI-H1869). (D) Binding sites between FGFRL1 and hsa-miR-107 were presented by bioinformatics. (E) Luciferase reporter assay was used to test the binding capacity of FGFRL1 and hsa-miR-107. (F) Overexpression efficiency of FGFRL1 was explored by qRT-PCR assays. (G) Luciferase reporter assay implemented to test the luciferase activity of wild and mutant FGD5-AS1. (H) Western blot and qRT-PCR assays were used to verify that interfered FGD5-AS1 and hsa-miR-107 can affect the expression of FGFRL1. **P<0.01 indicated statistically significant differences in this figure which n.s. indicated no significance.