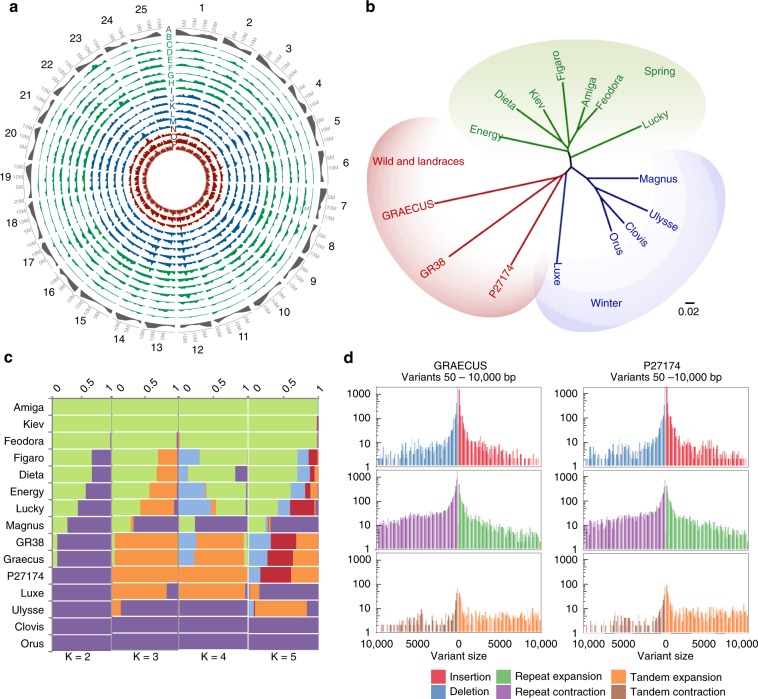
Fig. 2. White Lupin diversity and evolution history.
a SNP density identified by resequencing of 15 accessions of white lupin. In the outer track (A in gray) gene density is shown. Spring accessions are represented in green (B–H), winter accessions are represented in blue (I–M) and wild/landrace accessions are represented in red (N-P). From the outer to inner track: (B) AMIGA, (C) FEODORA, (D) KIEV, (E) DIETA, (F) FIGARO, (G) ENERGY, (H) LUCKY, (I) ORUS, (J) CLOVIS, (K) MAGNUS, (L) ULYSSE, (M) LUXE, (N) P27174, (O) GR38, AND (P) GRAECUS. The SNP density is represented in 1 Mb bins. b Neighbor-joining phylogenetic tree of white lupin accessions based on SNPs. The 15 accessions are divided in three clades: winter, spring and wild/landrace. c Admixture representation of the 15 accessions with population clustering for K = 2–5. Each individual is represented by a horizontal bar and each color represents a subpopulation. The color of each individual accession represents their proportional membership in the different populations. d Structural variants (SVs) between L. albus cv. AMIGA and the de novo assembly of GRAECUS (left) and P27174 (right). The biggest proportion of variants are the repeated elements. SVs represent 18.08 Mb of the GRAECUS genome and 18.67 Mb of P27174 genome. Source data underlying Fig. 2b are provided as a Source Data file.