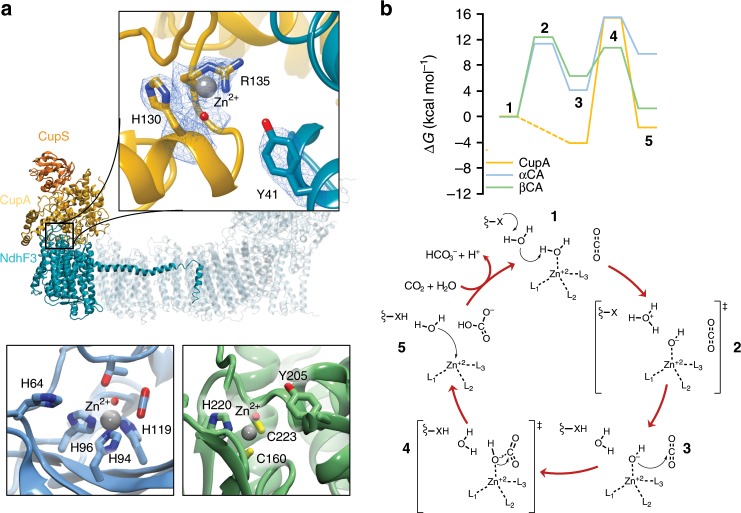
Fig. 2. Structural comparison and mechanism of carbon concentration in CupA and canonical carbonic anhydrases.
a The active site structure of the CO2 concentrating CupA subunit, showing the density of Zn-coordinating residues (5.5 sigma value, contour level 2, top), α-carbonic anhydrase (αCA, PDB ID: 5YUI, bottom left), and β-carbonic anhydrase (βCA, PDB ID: 1EKJ, bottom right). The Zn-bound density has been modelled as a water ligand, although the character of the ligand cannot be unambiguously assigned based on the map. b Reaction mechanism and free energy profiles for the CO2 hydration process based on quantum chemical DFT models in CupA, αCA, and βCA with Tyr41, His64, and Tyr205 as proton acceptors, respectively. Free energies are reported at the B3LYP-D3/def2-TZVP/def2-SVP/ε = 4 theory level (see Methods). Ligands L1/2/3 = His130/Arg135/H2O, L4 = H2O/OH−, X = Tyr41 in CupA; L1/2/3 = His94/96/119, L4 = H2O/OH−, X = His64 in αCA; L1/2/3 = Cys160/223/His220, L4 = H2O/OH−, X = Tyr205 in βCA. The CupA reaction takes place via two water molecules, modelled based on MD simulations (Supplementary Fig. 5j). See Supplementary Fig. 5 for further details.