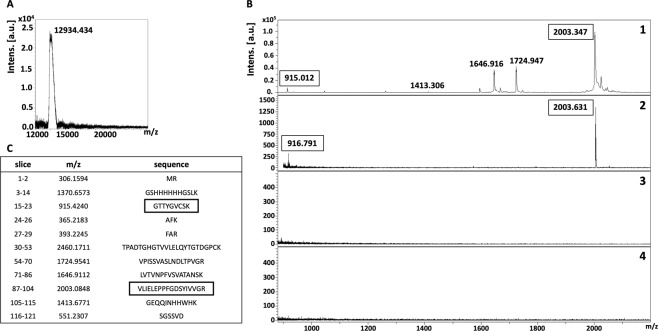
Figure 3.
Isolation of putative receptor-binding sites of rDIII using on-membrane limited tryptic digestion (nitrocellulose membrane). (A) (1) MALDI spectrum of in-solution limited tryptic digestion (20 min digestion) of rDIII, (2) rDIII incubated with ~15 kDa receptor on nitrocellulose membrane was trypsinized, interacting peptides were stripped, acetone precipitated and identified on MALDI-TOF, (3) negative control generated by omitting tryptic digestion, (4) negative control generated by omitting rDIII from the protocol. (B) Depicts rDIII recovered after interaction with ~15 kDa receptor from nitrocellulose membrane without trypsinization and detected on MALDI-TOF. C represents list of theoretical peptides of rDIII predicted by in-silico tryptic digestion using mMass software. Peptides identified from on-membrane limited proteolysis of ligand-receptor complex (A2), matching with in-solution digestion of ligand (A1) and theoretical masses (C) are framed. Please note that predicted masses of the peptides are [M + 1 H] +. The observed masses of the peptides are also [M + 1 H] +.