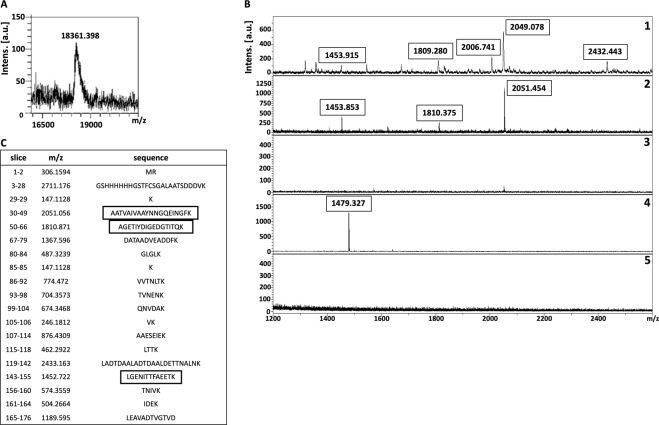
Figure 6.
Isolation of putative receptor-binding sites of rNadA using on-membrane limited tryptic digestion (PVDF membrane). (A) (1) MALDI spectrum of in-solution limited tryptic digestion (20 min digestion) of rNadA, (2) rNadA incubated with proteins of hBMECs immobilized on PVDF membrane was trypsinized, interacting peptides were recovered and identified on MALDI-TOF, (3) negative control generated by omitting tryptic digestion, (4) negative control generated by omitting rNadA from the protocol, (5) negative control generated by omitting proteins of hBMECs from the protocol. (B) Depicts rNadA recovered after interaction with proteins of hBMECs immobilized on PVDF membrane without trypsinization and identified on MALDI-TOF. C represents list of theoretical peptides of rNadA predicted by in-silico tryptic digestion using mMass software. Peptides identified from on-membrane limited proteolysis of ligand-receptor complex (A2), matching with in-solution digestion of ligand (A1) and theoretical masses (C) are framed. Please note that predicted masses of the peptides are [M + 1 H] +. The observed masses of the peptides are also [M + 1 H] +.