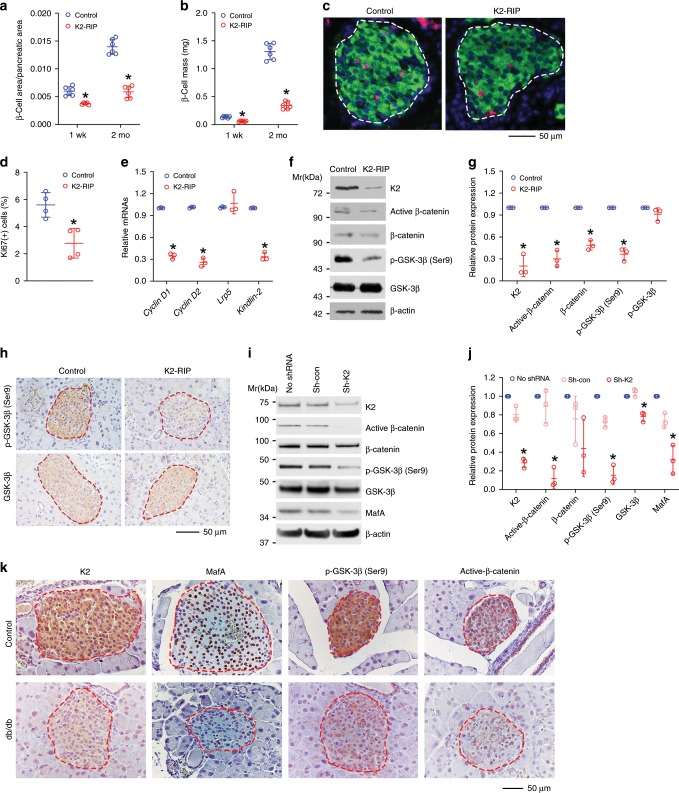
Fig. 3. Kindlin-2 loss activates GSK-3β and reduces β-catenin expression and β-cell proliferation and mass.
a, b Pancreatic sections from 1-week- or 2-month-old male control (RIP-Cre) and K2-RIP mice were subjected to IF staining for insulin, followed by measurements of β-cell area/pancreatic area ratio (a) and β-cell mass (b). *P < 0.05, versus control, N = 5 for 2-month-old K2-RIP in a, N = 6 for the remaining groups, Student’s t test. c, d Cell proliferation. Sections of 1-week-old control and mutant male pancreas were double stained with antibodies against insulin and Ki67 (c). Ki67-positive cells in islets were normalized to total insulin-positive cells in the same area (d). Around 444–816 β-cells (insulin-positive cells) per mouse were counted. N = 4 mice per genotype. Scale bar, 20 μm. *P < 0.05, versus control, Student’s t test. e qPCR analyses. Total RNAs isolated from islets of 1-week-old K2-RIP and control littermates were subjected to qPCR analysis for the indicated genes. mRNA levels were normalized to Gapdh mRNA. Statistical analyses (Student’s t test) were performed using the average values of triplicates from three independent experiments. *P < 0.05, versus control. f, g Western blot analysis. Protein extracts isolated from islets isolated from K2-RIP mice and control littermates (RIP-Cre) were subjected to Western blot analyses with indicated antibodies. Quantitative data from three independent experiments (g). *P < 0.05, versus control, Student’s t test. h IHC staining of pancreatic sections with antibodies against phospho-GKS-3β(Ser9) (top) and total GSK-3β (bottom). Scale bar, 50 μm. i, j Western blotting. Human pancreatic islets were infected with or without Kindlin-2 shRNA (Sh-K2) or control (Sh-con) lentiviruses, followed by Western blot analyses with indicated antibodies. Quantitative data from three independent experiments (j). *P < 0.05, versus control (Sh-con), Student’s t test. k IHC staining of pancreatic sections of 3-month-old db/db mice and age- and sex-matched C57BL/6 (control) mice with indicated antibodies. N = 3 mice per genotypes. Scale bar, 50 μm. Results are expressed as mean ± standard deviation. Source data for a, b, d–f, i, j are provided as a Source Data file.