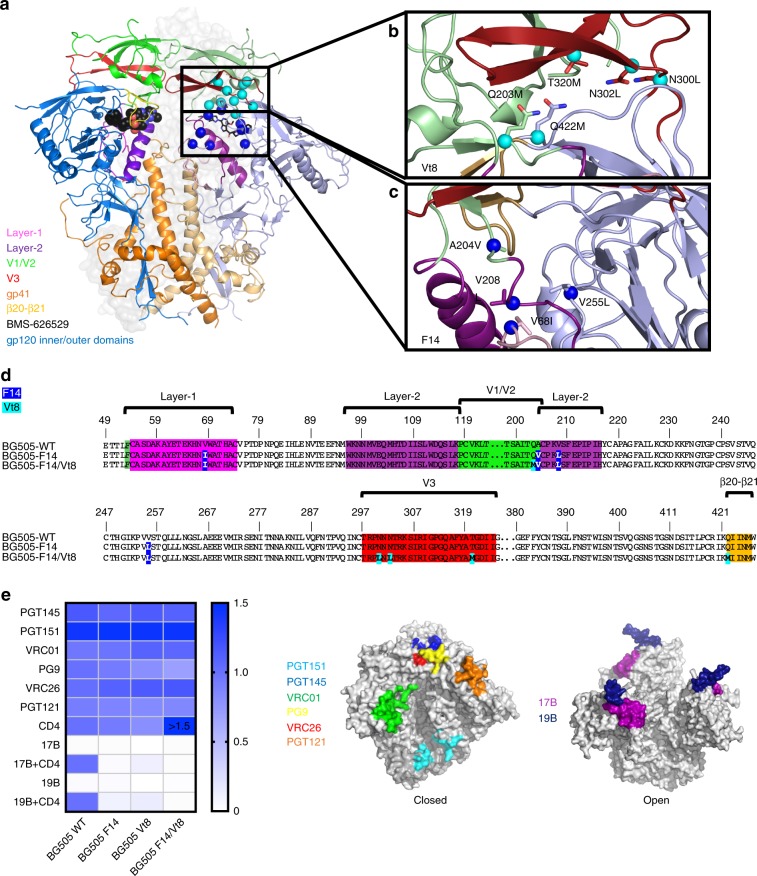
Fig. 1. Disrupting HIV-1 Env allostery to block CD4-induced conformational changes.
a HIV-1 Env SOSIP structure. Protomer to the left colored according to regions of allosteric control with BMS-626529 represented as black spheres. Protomer to the right in matte color scheme depicting the location of Vt-series mutations (cyan spheres) and F-series mutations (blue spheres). BMS-626529 depicted in stick representation. b Set of Vt8 residues. c Set of F14 mutations. Residue sidechains are represented as sticks with Cα atoms represented as cyan spheres. d Sequence alignment of BG505, F14, and F14/Vt8 with F14 mutations highlighted in blue and Vt8 mutations highlighted in cyan. Layers 1 and 2, the V1/V2 region, and β20-β21 are highlighted using the same color scheme as in a. e Binding antigenicity of BG505 SOSIP.664 and mutants. (left) Heatmap of bnAb binding responses, CD4 binding, and CD4 triggering. Values for CD4 binding and CD4 triggering are normalized to BG505 SOSIP. (middle) Surface representation of a closed state SOSIP (PDB ID 5CEZ) and (right) an open state structure (PDB ID 5VN3). Colors identify antibody binding locations.