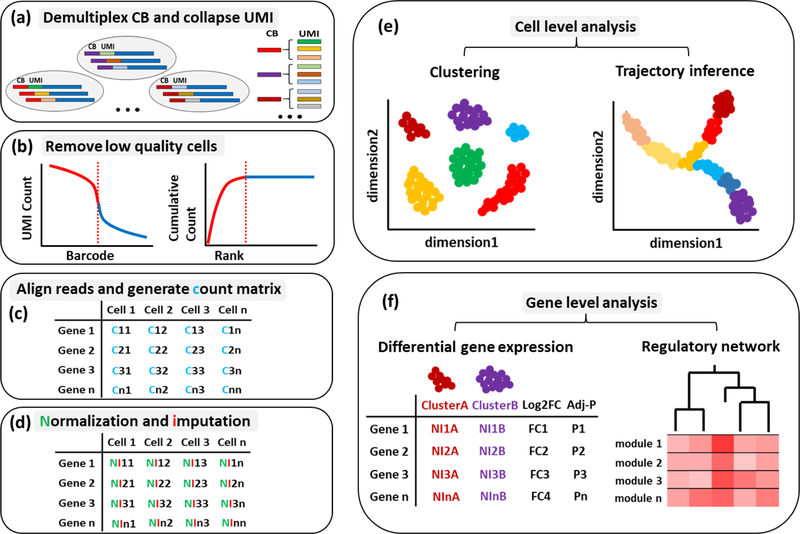
Figure 2. A schematic overview of scRNA-seq analysis workflow.
scRNA-seq data is noisy due to technical and biological factors. The analysis workflow involves (a) demultiplexing the sequencing reads by cell barcode and collapsing on UMI, (b) removing low quality cells (doublets, multiplets or dead cells), (c) aligning trimmed reads and generating count matrix, (d) normalization by scaling factors and imputation for sparse counts, (e) cell level analysis including clustering and trajectory inference and (f) gene level analysis including differential gene expression and regulatory network.