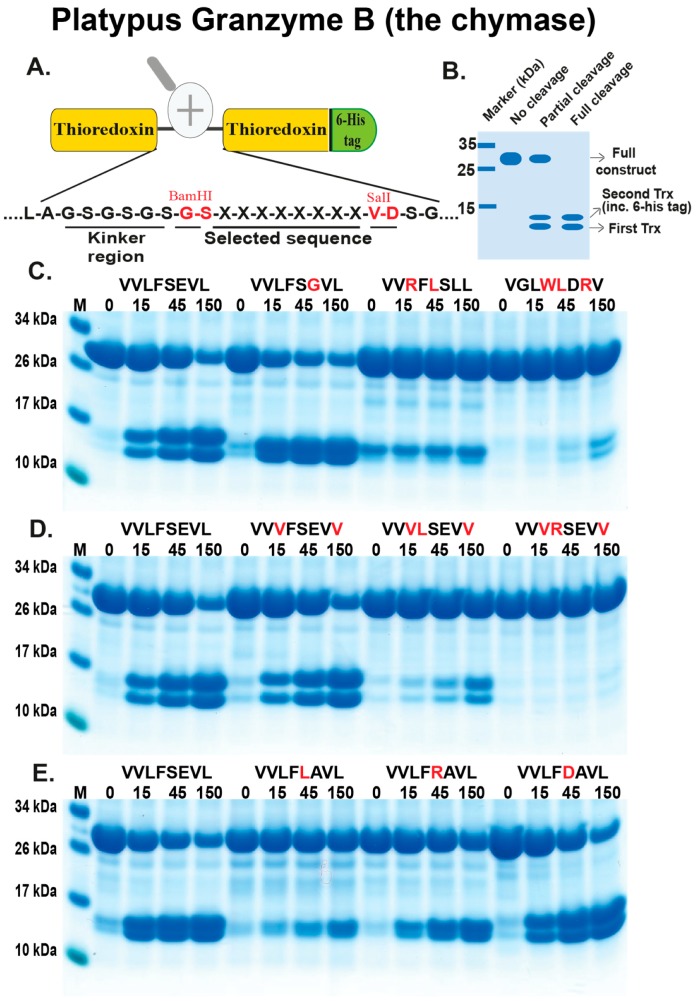
Figure 4.
Analysis of the cleavage specificity of platypus GzmB (the chymase) by the use of recombinant protein substrates. Panel (A) shows the overall structure of the recombinant protein substrates used for analysis of the efficiency in cleavage by the three different enzymes (analysed in Figure 4, Figure 5 and Figure 6). In these substrates, two thioredoxin molecules are positioned in tandem and the proteins have a His-6 tag positioned in their C-termini of the second thioredoxin. The different cleavable sequences are inserted in a linker region between the two thioredoxin molecules by the use of two unique restriction sites, one Bam HI and one SalI site, which are indicated in the bottom of panel (A). In panel (B), an example cleavage is shown to highlight possible cleavage patterns. Panels (C–E) show the cleavage of a number of substrates by the platypus granzyme B, the chymase. The sequences of the different substrates are indicated above the images of the gels. The red residues highlight the major difference between this substrate and the reference substrate positioned at the beginning (Left side) of this panel. The time of cleavage, in minutes, is also indicated above the corresponding lanes of the different gels. The un-cleaved substrates have a molecular weight of approximately 25 kDa and the cleaved substrates appear as two closely-located bands with a size of 12–13 kDa.