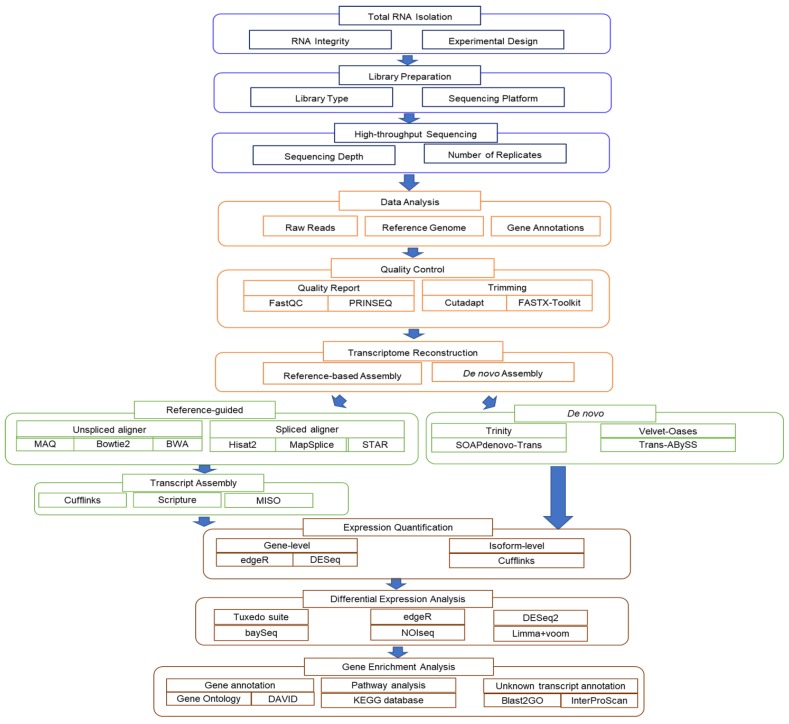
Figure 1.
General RNA-seq analysis pipeline. The workflow typically starts with total RNA extraction depending on experimental design and RNA integrity. The library preparation step relies on the selection of sequencing platform and library type, while sequencing depth and number of replicates can impact the downstream sequencing output analysis processes. RNA-seq data analysis generally requires inputs such as raw sequencing reads, reference genome sequences, and gene annotations. Next is examination of raw data quality and perform poor read trimming, transcriptome assembly, and expression quantification. Finally, differential expressed genes (DEGs) must be identified and interpreted through gene enrichment analysis. Each step in the data analysis has several representative tools, as highlighted.