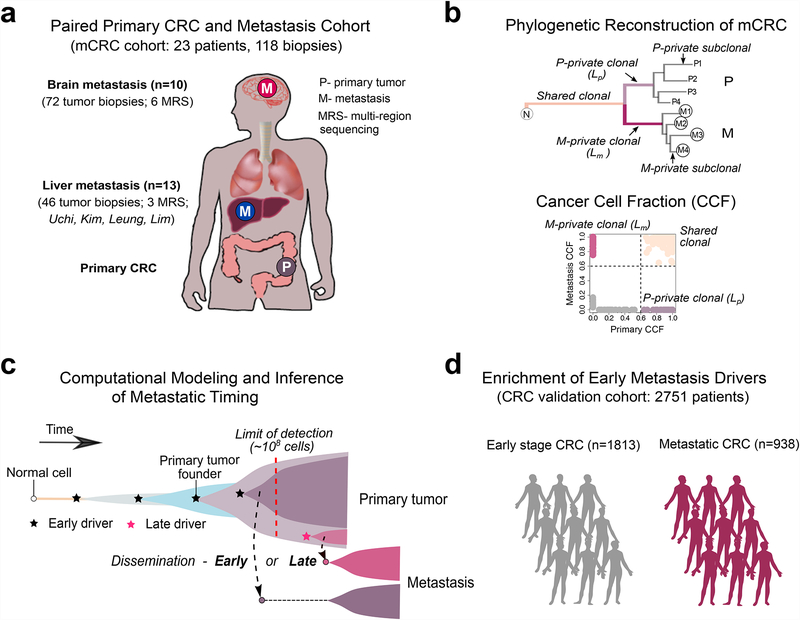
Figure 1. Study overview.
(a) The metastatic colorectal cancer (mCRC) patient cohort includes 118 tumor biopsies from 23 patients. Paired CRCs with metastases to the brain and other sites (liver, lung, lymph nodes) from 10 patients and 72 tumor biopsies were whole-exome sequenced, including 6 cases with multi-region sequencing (MRS) of 3–5 regions each from the primary CRC and metastasis. Additionally, four publicly available cohorts with paired CRCs and liver metastases from 13 patients and 46 tumor biopsies were reanalyzed within the same bioinformatics framework, including 3 cases with MRS. (b) Tumor phylogenies were reconstructed from somatic alterations (sSNVs+indels). The mutational cancer cell fraction (CCF) was computed and compared for each primary CRCs and metastasis pair. (c) Schematic illustration of tumor evolution starting from a normal cell that acquires mutations leading to malignant transformation, growth of the primary tumor, metastatic dissemination, seeding and outgrowth. It is unknown whether dissemination occurs early from a dominant subclone when the size of the primary tumor is below the limits of clinical detection (108 cells or 1 cm3) (early dissemination) or later from a minor subclone after the acquisition of additional driver alterations (late dissemination). To address this question, we developed a 3-D model of tumor growth and statistical inference framework to time metastasis from patient genomic data. (d) We further leveraged a large collection of metastatic (n=938) and non-metastatic (n=1,813) CRCs with targeted sequencing data to evaluate the association between specific combinations of early driver genes (modules) identified in the mCRC cohort.