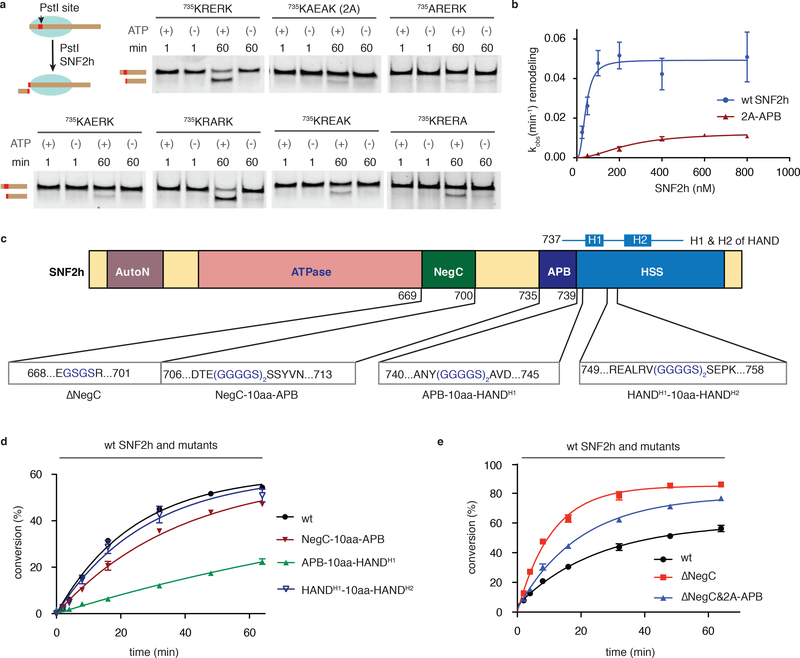
Figure 3 |. The ABP motif is required for SNF2h-mediated nucleosome remodeling.
(a) Remodeling activity of wild-type SNF2h and APB mutants (indicated in bold) was assessed by REAA. Schematic of the assay shown at top right; PstI = restriction endonuclease. Reactions (± ATP) were analyzed at indicated times by native polyacrylamide gel with SYBR® gold staining. Uncropped figures are shown in Supplementary Fig. 5b. (n=3 independent experiments). (b) Plot of observed remodeling rates, as measured by REAA, versus SNF2h concentration for both wild-type and the 2A-APB mutant. Data were fit to a Hill slope model yielding Kmapp values of 43 ± 7 nM and 243 ± 56 nM for wt SNF2h and 2A-APB, respectively. Errors = s.e.m (n=3 independent experiments). (c) Schematic showing deletion and insertion mutants in SNF2h designed to explore the interplay between the APB and the flanking NegC and HSS domains. The H1 and H2 helices in the HAND domain are indicated above. (d,e) Kinetics of remodeling as measured by REAA for the indicated SNF2h mutants relative to wild-type. Errors = s.e.m (n=3 independent experiments). For b,d and e data are represented as the mean of experimental replicates.