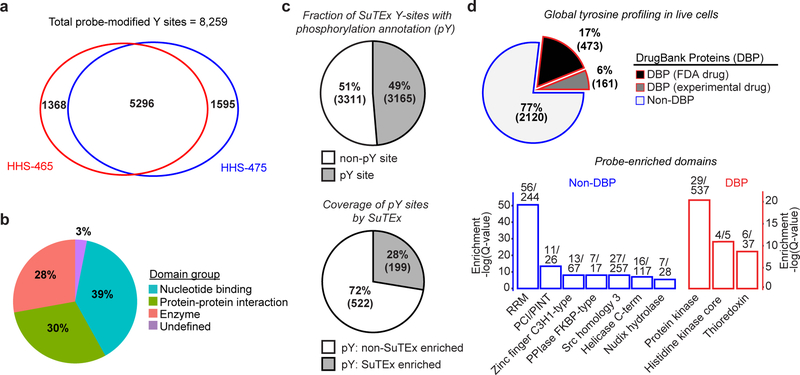
Figure 2. Functional tyrosine profiling in proteomes and live cells.
a) Comparison of HHS-465- and HHS-475-tyrosine modified sites identified from human cell proteomes (HEK293T, A549, DM93, H82, and Jurkat cells) treated with SuTEx probes (100 μM, 1 hr, 25 °C). b) Distribution of protein domain groups that are significantly overrepresented using probe-modified tyrosine sites from in situ chemical proteomic studies. Enriched domain annotations are those with a Q-value < 0.01 after Benjamini–Hochberg correction of a two-sided binomial test (see Online Methods for details). c) Top: Overlap between in situ HHS-465- and HHS-475-modified tyrosine sites that are also phosphorylation sites (number of phosphotyrosine high throughput annotation on PhosphoSitePlus (HTP score); HTP ≥1). Bottom: coverage of phospho-tyrosine sites (HTP ≥10) that were detected by in situ chemical proteomics of HEK293T and Jurkat cells (HHS-465 and −475). d) Top: Comparison of HHS-465 and HHS-475 in situ probe-modified proteins with DrugBank proteins (DBP group). The Non-DBP group consists of proteins that did not match a DrugBank entry. Bottom: probe-enriched domains from DBP and non-DBP groups. Enriched domain annotations are those with a Q-value < 0.01 after Benjamini–Hochberg correction of a two-sided binomial test. All data shown are representative of two experiments (n=2 biologically independent experiments).