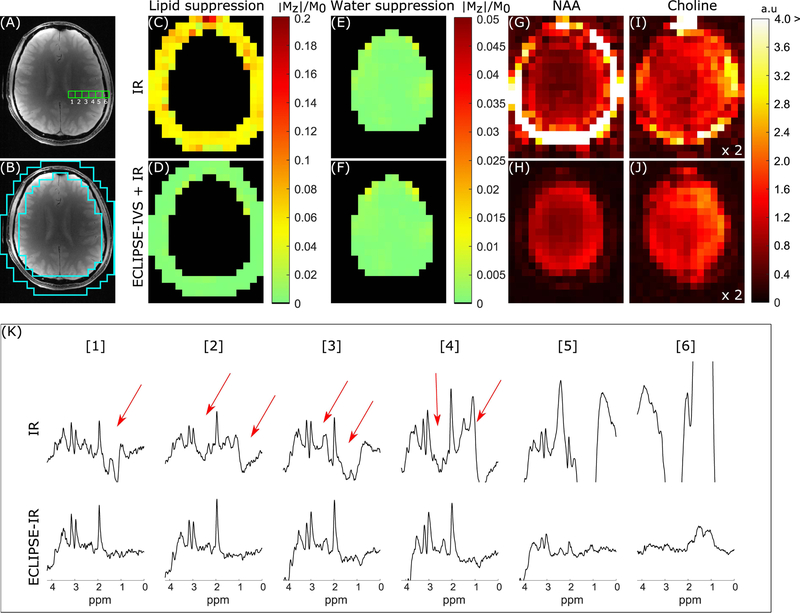
FIG. 5.
Summary of IR and ECLIPSE-IVS + IR sequences with identical layout to Figure 4. (A) The anatomical image corresponding to the MRSI slice illustrating six adjacent MRS voxel locations used to compare the two IR methods in (K). (B) The ROI used to calculate extracranial lipid suppression in (C–D). (C–D) Residual extracranial lipid fraction maps for the IR and ECLIPSE-IVS + IR sequences, respectively. (C) Near-uniform lipid suppression throughout the ROI is seen (relative to conventional-OVS) with ~18-fold mean suppression with IR (mean/median |Mz|/M0 of 0.057/0.054, respectively), and (D) ECLIPSE-IVS + IR in comparison, demonstrates superior lipid suppression with a mean suppression factor of ~257-fold (mean/median |Mz|/M0 of 0.0039/0.0025, respectively) with no hotspots. (E–F) Residual water fraction maps within the brain for IR and ECLIPSE-IVS + IR sequences with near identical mean water suppression performance of ~760-fold (mean/median |Mz|/M0 of 0.0013/0.0009, respectively). (G–H) NAA and (I–J) Choline maps calculated for the two methods. NAA maps with Global IR is dominated by extracranial lipids, which continues to bleed into choline maps, while no such hyper-intensities are seen with ECLIPSE-IVS + IR. The less pronounced ventricular outline in this case (relative to that of Figure 4) is primarily due to the low level of CSF in the slice. Note that the axial slices in Figure 4 and Figure 5 are from different subjects and slice positions. (K) Spectra from the 6 adjacent voxels shown in (A). Voxels 1–4 with IR show prominent metabolites (NAA, creatine, and choline) with significant lipid contaminations (marked with red arrows). In comparison minimal contaminants are observed from voxels 1–4 with ECLIPSE-IVS + IR, and lipid contaminants in voxel 6 is seen to be below the amplitude of NAA in brain, but are beyond displayed bounds for IR.