Abstract
Genetically encoded fluorescent proteins have been used for metal ion detections by combining fluorescent proteins with metal-binding proteins or peptides. However, their applications are largely restricted to a limited number of metal ions, such as Ca2+ and Zn2+, due to the lack of available metal-binding proteins or peptides that can be fused to fluorescent proteins and the difficulty in transforming the binding of metal ions into a change of fluorescent signal. To overcome these limitations, we report herein the use of Mg2+-specific 10-23 or Zn2+-specific 8-17 RNA-cleaving DNAzymes to regulate the expression of fluorescent proteins as a new class of ratiometric fluorescent sensors for metal ions. Specifically, we demonstrate the use of DNAzymes to suppress the expression of Clover2, a variant of the green fluorescent protein, by cleaving the mRNA of Clover2, while the expression of Ruby2, a mutant of the red fluorescent protein, is not affected. The Mg2+ or Zn2+ in Hela cells can be detected using both fluorescent confocal imaging and flow cytometry. Since a wide variety of metal-specific DNAzymes, such as for Mg2+, Na+, Cu2+, Zn2+, Pb2+, Hg2+, Ag+, and UO22+, can be obtained through in vitro selection, and the resulting DNAzymes often share a similar secondary structure and reaction mechanism, the method described in this work can likely be applied to imaging many other metal ions and thus significantly expand beyond the range of the current genetically-encoded fluorescent proteins, allowing this class of sensors to be even more powerful in providing deeper understanding of the roles of metal ions in biology.
Keywords: DNAzyme, Fluorescent Proteins, Genetically Encoded Sensor, Metal ions
Graphical Abstract
Using metal-specific RNA-cleaving DNAzymes to regulate the expression of fluorescent proteins has expanded the genetically-encoded fluorescent protein-based sensors for metal ions significantly.
Introduction
Metal ions play crucial structural and functional roles in biological processes. Understanding the distribution and concentration fluctuation of metal ions in cells is a central topic in the bioanalytical and biomedical sciences with many potential impacts, from cell signaling to metabolic engineering to medical diagnostics and imaging.[1] Towards this goal, fluorescent sensors for cellular metal ions have been developed based on a variety of molecules, including small organic molecules,[2] polymers,[3] and proteins.[4] Among them, genetically encoded fluorescent sensors,[4a, 4c-l] pioneered by Tsien and co-workers,[4a] have been a major focus by numerous groups around the world for many years, because they can be generated by fusing fluorescent proteins to metal-binding proteins or peptides. The resulting fusion proteins often preserve the biochemical functions and cellular localization of the partner proteins. These sensors have been applied for intracellular metal ion detection, especially in monitoring the homeostasis of subcellular organelles, such as mitochondrion,[5] endoplasmic reticulum, and Golgi apparatus.[4h] While much progress has been made, this class of sensors can detect only a limited number of metal ions, such as Ca2+, Mg2+, and Zn2+, due to the lack of available metal-binding proteins or peptides that can be fused to fluorescent proteins and the difficulty in transforming the binding of metal ions into a change in the fluorescent signal. For example, to extend this class of sensors to detecting Mg2+ in cells,[4i, 6] a number of mutations were made to adapt a truncated version of the HsCen3 protein, which contains two EF-hand metal-binding sites that normally bind Ca2+ strongly, to also bind Mg2+ with a dissociation constant (Kd=148 μM) around 10 fold weaker than that of Ca2+ (Kd=10 μM). In addition, to transform the binding of Mg2+ by this mutant protein into genetically encoded fluorescent protein signals, further engineering of the protein construct was required in order to trigger enough conformational change of the protein to generate a detectable shift in the Főrster Resonance Energy Transfer (FRET) fluorescent signal. Because of the difficulty and complexity in transforming metal-binding proteins or peptides into fluorescent sensors through large conformational changes, this method has been limited to sensing only a few metal ions. Therefore, the method of using genetically encoded fluorescent sensors to detect a wide number of other metal ions remains a challenge and thus has not reached its full potential.
To meet such a challenge, we examined the possibility that RNA-cleaving DNAzymes could be used in conjunction with genetically encoded fluorescent proteins for metal ion detection and ratiometric imaging of metal ions in living systems. DNAzymes are catalytic DNA molecules that can perform an enzymatic function, often in the presence of specific metal ions that act as cofactors in the catalysis.[7] DNAzymes are obtained through in vitro selection from a large library of 1015 different sequences, which is subjected to several rounds of selection pressure in order to separate out and identify those sequences that undergo the desired chemical reaction under the desired conditions, such as in the presence of a specific metal ion.[7a, 8] For the case of metal-specific DNAzymes, an RNA cleavage reaction is typically selected in which the DNAzyme can site-selectively cleave an RNA bond within a specific substrate strand. Furthermore, selectivity for the desired metal ion can be significantly enhanced by performing negative selections against other similar ions that could have competitive binding, in order to remove any sequences that react with these metal ions and keep only those sequences that react with the metal ion of interest. Using these techniques, many different DNAzymes have been selected that have high specificity for many different biologically relevant metal ions. As a result, many fluorescent sensors have been constructed based on these DNAzymes to detect these metal ions, including Mg2+, Zn2+, UO22+, Pb2+, Ag+, and Na+, in either environmental or biological samples, by coupling the metal-selective cleavage of an RNA-containing substrate into a fluorescent signal using the catalytic beacon approach.[7f, 8c, 9] Despite the potential of these DNAzymes as metal ion sensors, they have been applied to detecting cellular metal ions only within the past six years and in limited cases.
In order to expand upon the number of metal ions that the genetically encoded ratiometric fluorescent protein sensors can detect and to increase the impact of DNAzyme sensors in cellular biology, we herein report the use of the 10-23 DNAzyme to regulate the expression of fluorescent proteins as a method to detect intercellular Mg2+. To demonstrate the generality of this approach, we have also used the same approach with the 8-17 DNAzyme to ratiometrically image Zn2+ in living cells with genetically expressed plasmids.
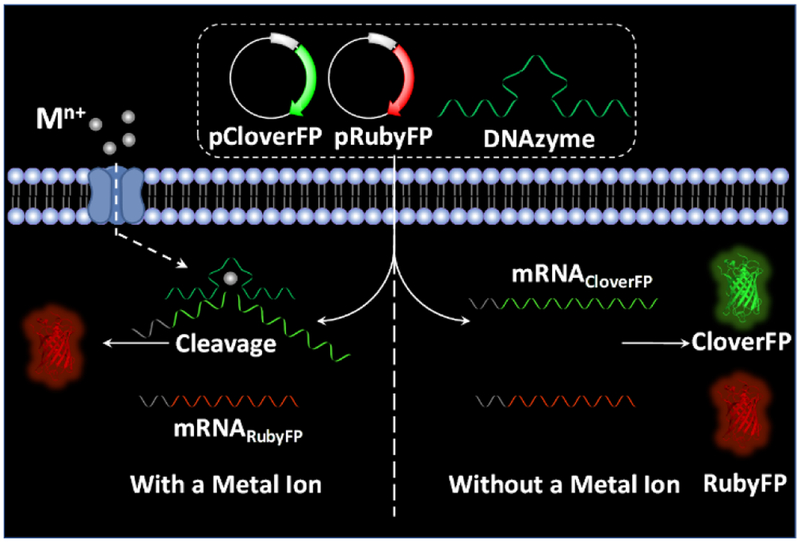
Results and Discussion
Fluorescent proteins show many advantages when employed as a fluorescent signal reporter. In addition to the biocompatibility and photostability, highly expressed fluorescent proteins will generate a strong fluorescent signal, which is crucial for successful cell imaging. In our design, we chose Clover2 and mRuby2,[10] mutants of green and red fluorescent proteins, as a signal reporter and a permanent reference, respectively, for ratiometric measurement, and name them CloverFP and RubyFP. To couple the fluorescent signal changes from CloverFP/RubyFP with metal-specific DNAzymes, we use a DNAzyme to suppress the expression of CloverFP in the presence of the metal ion of interest, while the expression of RubyFP will not be affected by the same DNAzyme. As shown in Scheme 1, the DNAzyme and two plasmids expressing CloverFP and RubyFP (pCloverFP and pRubyFP, respectively), can be co-transfected into HeLa cells. The plasmids could further be transcribed into mRNA transcripts for CloverFP and RubyFP (called mRNACloverFP and mRNARubyFP, respectively). As the arms of the DNAzyme can be designed to hybridize with a specific location within mRNACloverFP, the mRNACloverFP would be cleaved by the DNAzyme in the present of its metal ion, while the mRNARubyFP would not be cleaved because it would not contain this DNAzyme hybridization sequence. Therefore, the cells would express much more RubyFP than CloverFP, of which the fluorescent signal can be observed by either confocal microscopy imaging or flow cytometry. In the absence of the metal ion that the DNAzyme uses for mRNA cleavage, the ratio of the RubyFP fluorescence to CloverFP fluorescence (RRubyFP/CloverFP) will be close to one, since they are both under the same promoter and so should have similar expression levels. In the presence of the metal ion, the DNAzyme would cleave mRNACloverFP, suppressing the CloverFP expression and resulting in an increased RRubyFP/CloverFP.
Scheme 1.
Schematic illustration of DNAzyme-mediated genetically encoded sensors for ratiometric imaging of metal ions in living cells.
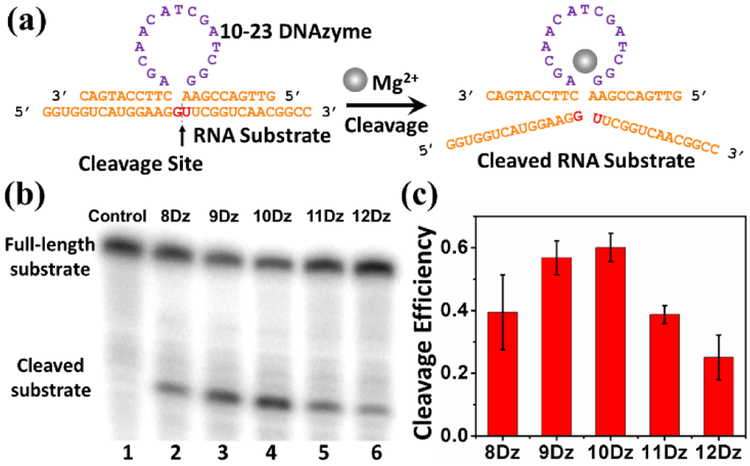
RNA-cleaving DNAzymes have been reported to cleave full RNA strands, which provide a potential alternative for the regulation of gene expression.[11] In 2010, Deiters and co-workers reported a DNAzyme-based ratiometric sensor that hybridized to the target mRNA of a fluorescent protein through the binding arms of DNAzymes, which induced degradation of the mRNA.[12] By using binding arms of both inactive DNAzymes and DNA strands that didn’t contain the DNAzyme catalytic core, they also found similar levels of mRNA degradation and thus concluded that their DNAzyme methodology in fact achieved the majority of its mRNA silencing not through its DNAzyme-mediated mRNA cleavage activity, but rather via a typical antisense mechanism which utilizes cellular RNAse H to recognize RNA: DNA duplexes to degrade target mRNA. As a result, this previous design cannot be used for sensing metal ions, because the antisense function is not dependent on Mg2+ concentration. We overcome this issue with careful optimization of DNAzyme sensor design, mRNA target site selection, and cellular transfection conditions to allow for optimal DNAzyme cleavage activity of our intended mRNA target, as well as the use of inactive DNAzyme negative controls, to ensure that signal response is due to the Mg2+-dependent multiple turnover activity of DNAzymes, rather than other cellular degradation pathways such as RNAse H. We chose the 10-23 DNAzyme for this study because it has been reported to cleave mRNA both in vitro and in vivo.[13] As shown in Figure 1a, the 10-23 DNAzyme strand consists of a 15-nucleotide (nt) single-stranded DNA region called the catalytic domain (purple), which is critical to the DNAzyme cleavage activity. Two binding arms (orange) flanking the catalytic domain can hybridize to an RNA substrate strand containing the cleavage site (red) which is directly opposite to the catalytic core (Figure 1a). In the cleavage site, it has been determined that the susceptibility of the dinucleotide influences the DNAzyme activity, with the trend being AU = GU ≥ GC >> AC.[14] Therefore, we have selected the GU dinucleotide pair for this study. In addition to the cleavage site, the length of the two binding arms plays a role in determining the effectiveness of hybridization before cleavage and release of the product after the cleavage,[15] which is critical for catalytic turnover of the DNAzymes. Therefore, the 10-23 DNAzyme with 8-, 9-, 10-, 11- or 12-nt in the binding arms, called 8Dz, 9Dz, 10Dz, 11Dz, 12Dz, respectively, were designed to investigate their effect on the reactivity. (See Table S1 in ESI for complete sequence information).
Figure 1.
(a) The structure of the 10-23 DNAzyme and detection of the RNA substrate cleavage catalysis. (b) Gel electrophoresis analysis of the RNA cleavage efficiency by using 8Dz, 9Dz, 10Dz, 11Dz, and 12Dz with a DNAzyme: substrate ratio of 10:1. (c) The quantification of the gel electrophoresis analysis.
To assess the activity of the above DNAzyme constructs, the RNA substrate was labelled with γ-32P ATP before the 10-23 DNAzymes were added in the presence of 30 mM Mg2+ in Tris-HCl buffer. After 60 minutes, the solution was quenched by stop-solution and loaded onto denaturing polyacrylamide gel electrophoresis (PAGE) to determine the percentage of the cleaved product from the RNA substrate. To investigate the ability of multi-turnover cleavage by the DNAzyme, an excess amount of substrate was used to make the ratio of DNAzyme: substrate to be 1:10. As shown in Figure 1b, obvious lower band, likely representing the cleaved substrate product can be observed for all DNAzyme lengths tested, from lane 2 to lane 6, after the addition of the DNAzymes. In contrast, no cleaved product band was found in the absence of DNAzyme (lane 1), which demonstrates that all of the DNAzymes with different lengths of binding arms were able to cleave the RNA substrate. Quantitative analysis of the results indicates that the DNAzymes exhibited cleavage efficiency higher than 0.1 (the maximum possible efficiency for single-turnover reaction under a 1:10 enzyme: substrate ratio), which strongly suggests that the DNAzymes carry out more than one cleavage reaction turnover (Figure 1c). Among these tested DNAzymes, the 10-23 DNAzyme with 10-nt binding arms (10Dz) showed the highest multi-turnover cleavage efficiency, which is consistent with results reported previously,[16] as the shorter binding arms cannot form an efficient hybridization with the RNA substrate, while the longer binding arms could inhibit the release of the cleaved substrate. Additionally, inactive DNAzymes that have a point mutation from G to C within the catalytic domain (named 8DzM, 9DzM, 10DzM respectively) were tested as negative controls to rule out any possible artifacts in target RNA cleavage that are independent of Mg2+-based activity. As the results show in Figure S2, the RNA substrate was efficiently cleaved in the active DNAzyme constructs, while the mutation in the catalytic domain led to almost no catalytic activity, which indicates that the cleavage of RNA substrate was indeed caused by the catalytic function of the DNAzyme. Finally, the cleavage efficiency of 10Dz towards the RNA substrate under different concentrations of metal ions was also studied. As displayed in Figure S3, the 10Dz displayed higher enzymatic activities in the presence of increasing concentrations of Mg2+. Therefore, 10Dz, the 10-23 DNAzyme with flanking arms of 10 bases, was selected for further experiments.
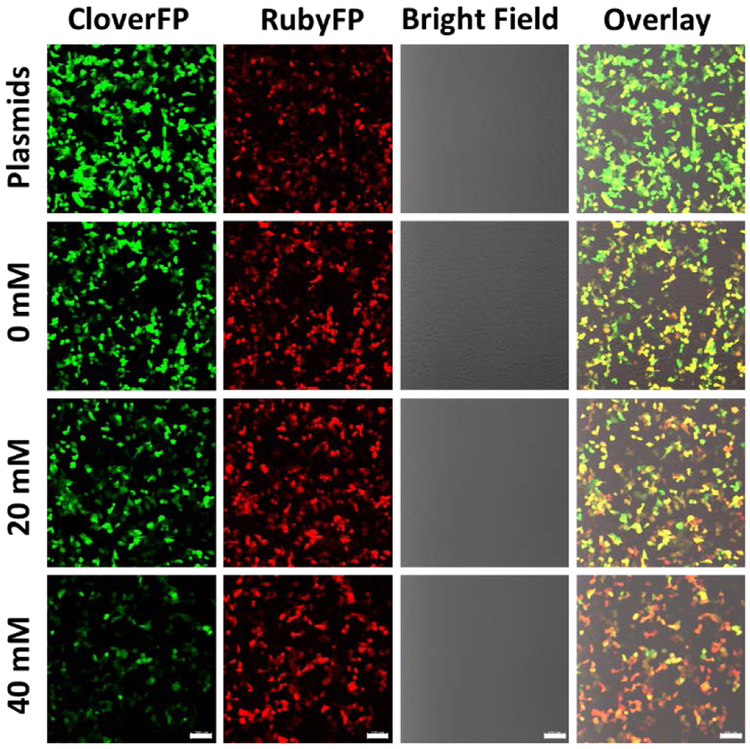
In order to investigate whether the above 10-23 DNAzyme construct can be used to specifically cleave the mRNA coded for a fluorescent protein in mammalian cells, we searched GU sites in mRNACloverFP (see complete sequence of mRNACloverFP in ESI). Based on minimal free energy predictions of the secondary structures of mRNACloverFP in Figure S1, we identified GU at positions 49-50 to be an accessible region for cleavage by the DNAzyme. To protect the DNAzymes from degradation by exonucleases in cells, the DNAzymes were modified with either inverted T at 3´ end (Dz10TCloverFP) or two hairpins at both the 3´ and 5´ ends (Dz10HCloverFP). Either Dz10TCloverFP or Dz10HCloverFP was co-transfected in HeLa cells with both pCloverFP as the reporter and pRubyFP as the reference for ratiometric imaging. To find the optimal conditions for plasmid expression and DNAzyme transfection, the time-dependent activity of DNAzymes in cells was studied by confocal imaging (Figure S4). After 24 hours of total incubation (transfection and Mg2+ treatment), the cells transfected with Dz10TCloverFP or Dz10HCloverFP showed a higher ratiometric signal than the cells transfected with plasmids only, without the DNAzymes. Extending the total measurement time to 48 hours could not significantly increase the ratiometric change of the sensor, and the change was decreased after 72 hours of total incubation. This decrease of sensor performance during longer incubation could be explained by the degradation of the DNAzymes under cellular conditions. Based on these observations, we decided to use the protocol of 6 hours transfection with 18 hours ion treatment (24 hours total) before fluorescence quantification by confocal microscopy imaging or flow cytometry for later experiments. After 6 hours of incubation, different concentrations of Mg2+ were delivered into cells with the assistance of 2 μM calcimycin, which was demonstrated to show no interference in the DNAzyme reactivity (Figure S5). The cells were subsequently incubated for another 18 hours to allow the expression and maturation of the fluorescent proteins. The transfection efficiency of plasmids and DNAzymes were measured by flow cytometry. The expression efficiency of CloverFP and RubyFP were observed to be 57.1% and 57.3%, indicating a stable co-transfection of the plasmids, and the transfection efficiency of Dz10TCloverFP and Dz10HCloverFP were observed to be 95.6% and 93.6%, respectively (Figure S6). The confocal microscopy imaging demonstrated that when Mg2+ was applied into cells after Dz10TCloverFP and Dz10HCloverFP transfection, the fluorescent signal in the green channel where CloverFP emits decreased, while the red channel where RubyFP emits remained relatively constant (Figure 2 and Figure S7). Furthermore, the change of ratios between two fluorescent proteins can also be observed visually by examining the changes in the “overlay” merged channel (Figure 2, Figure S7) and the RubyFP/CloverFP ratio channel of single cells (Figure S8) with increasing concentrations of Mg2+. These results suggest our design in Figure 1 works toward imaging metal ions in living cells.
Figure 2.
Confocal imaging of the cells expressing CloverFP (green channel) and RubyFP (red channel). The cells were transfected with plasmids only (first row) or with plasmids and Dz10TCloverFP with different concentrations of added Mg2+ (second to fourth rows). Scale bar = 100 μm.
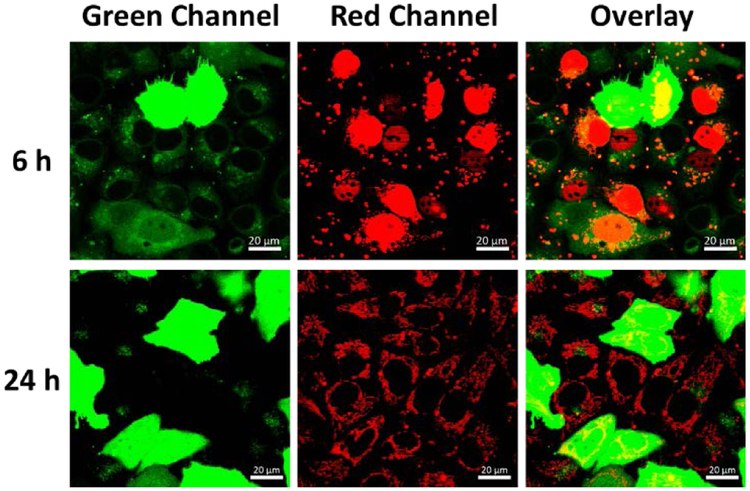
To better understand the pathway of DNAzymes after co-transfection with the plasmids, a Cy5 labelled DNA (see Table S1 in ESI for sequence information) was co-transfected with the CloverFP plasmid and the localization of the Cy5-DNA was tracked after 6h and 24h (Figure 3). Since the CloveFP needs a relatively long time to express and mature, the lysosome stained by LysoGreen can still be visualized at 6h. The absence of co-localization between the Cy5-DNA (red channel) and the lysotracker (green channel at 6h) indicated that most of the Cy5-DNA resides outside of lysosomes. Interestingly, the Cy5-DNA was found to be localized largely to the nucleus at 6h transfection time, which might be caused by the transfection reagent.[17] However, by 24h transfection time, the Cy5-DNA is dispersed throughout the cytoplasm.
Figure 3.
Confocal imaging of cells co-transfected with CloverFP plasmid (green channel) and Cy5-DNA (red channel) at 6 h and 24h. The lysosomes of cells were stained with Lysotracker Green. (The CloverFP expressing cells exhibited green fluorescence in the whole cells). Scale bar = 20 μm.
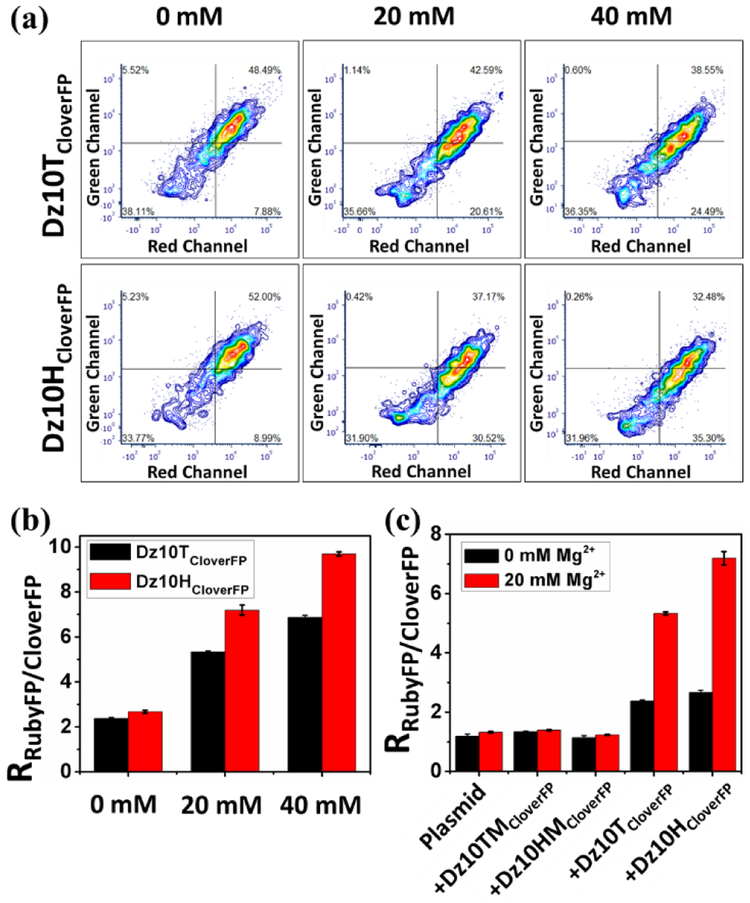
To ensure the results from confocal microscopy imaging apply to the entire cell population, we examined the fluorescence in a large cell population using flow cytometry. Two channels were used to measure the fluorescence intensities. As shown in Figure 4a, both Dz10TCloveFP and Dz10HCloverFP induced a fluorescence decrease in the green channel in the presence of Mg2+, while no obvious change was observed in the red channel. Correspondingly, the ratio of RubyFP to CloverFP increased when more Mg2+ was introduced into the cells, suggesting a direct correlation between this ratio, as an indicator of DNAzyme activity, and Mg2+ concentration (Figure 4b). This result is in strong agreement with the confocal imaging results, suggesting the successful detection of Mg2+ through this approach. To further demonstrate that this signal change was caused by the catalysis of DNAzymes rather than RNase H mediated RNA degradation, the mutant DNAzymes Dz10TMCloverFP and Dz10HMCloveFP were also employed as negative controls. As shown in Figure 4c, minimal change of the ratio was detected by the inactive Dz10TMCloverFP and Dz10HMCloverFP over the plasmid control with or without addition of 20mM Mg2+. In contrast, the ratio was significantly increased when the active Dz10TCloverFP or Dz10HCloverFP was used, which implies that the downregulation of the mRNACloverFP is induced by the catalysis of the active DNAzymes. Moreover, even without additional Mg2+, both active DNAzyme constructs (Dz10TCloverFP and Dz10HCloverFP) showed higher ratios than the mutant DNAzymes (Dz10TCloverFP and Dz10HCloverFP), and this ratio increased even further when 20 mM Mg2+ was added. These results suggest that the active DNAzyme constructs can detect endogenous Mg2+ in cells and the ratios depend on Mg2+ concentrations. In order to obtain even better quantification of the metal ions in the cells, DNAzymes with affinity for metal ions in their physiological concentration range needs to be obtained, either through optimization of the sensor design or re-selection of DNAzymes.[18]
Figure 4.
(a) The contour charts of flow cytometry when Dz10TCloverFP or Dz10HCloverFP was applied to different concentrations of Mg2+. (b) Fluorescent signal ratio of RubyFP to CloverFP in different concentrations of Mg2+ when Dz10TCloverFP or Dz10HCloverFP was used. (c) Comparison of the ratio generated by Dz10TCloverFP or Dz10HCloverFP and Dz10TMCloverFP or Dz10HMCloverFP in the absence or presence of Mg2+.
To investigate the generality of our ratiometric ion detection method to be applied to other DNAzymes for detection of different metal ions in living cells, the 8-17 DNAzyme, a Zn2+-specific RNA-cleaving DNAzyme, was employed to construct another genetically encoded ratiometric sensor using the same strategy. The 8-17 DNAzyme has been demonstrated to cleave full RNA sequences at a GG dinucleotide junction.[19] The activity of the 8-17 DNAzyme (10Zn-GG) version of the ratiometric sensor was confirmed by polyacrylamide gel electrophoresis (Figure S9).
More importantly, after screening GG junctions in both mRNARubyFP and mRNACloverFP (see Figure S1 for the sequences investigated) by ratiometric imaging, we found that 10ZnC targeting the mRNACloverFP was able to downregulate the CloverFP under endogenous Zn2+ levels (Figure S10). Moreover, quantifying the RRubyFP/CloverFP ratio showed a further increase after introducing additional Zn2+ in cell culture. Therefore, our strategy can be generalized and applied to image intracellular Zn2+ (Figure S11) in addition to intracellular Mg2+.
Conclusion
In summary, we have successfully developed a novel fluorescent imaging method for ratiometric imaging Mg2+ and Zn2+ in living cells based on DNAzyme-mediated genetically encoded fluorescent proteins, which to our knowledge, is the first demonstration of DNAzyme-mediated genetically encoded sensors for intracellular detection of metal ions. Even though numerous fluorescent proteins have been reported for more than 30 years for other targets, few sensors have been applied to imaging metal ions in living cells. In comparison to previous publications in which the mRNA of fluorescent proteins was mainly degraded by the hybridization of DNAzymes with mRNA to induce degradation through a classical antisense mechanism using RNAse H rather than DNAzyme-mediated RNA cleavage,[12] our method possesses the advantage of Mg2+-dependent multi-turnover cleavage toward target mRNA by the activity of DNAzymes. This advantage leads to a correlation between the expression levels of fluorescent protein and the concentration of target metal ions. By using Mg2+- or Zn2+-specific DNAzymes to mediate expression of fluorescent proteins, we demonstrate the use of fluorescent proteins to image Mg2+ and Zn2+ in living cells. Having demonstrated that this methodology can be applied to two different DNAzymes, it is likely that it can be further applied to other RNA-cleaving DNAzymes, as long as they are capable of cleaving an all-RNA substrate. Thus, in utilizing the many different metal-specific DNAzymes for metals, such as Mg2+, Na+, Cu2+, Zn2+, Pb2+, Hg2+, Ag+, and UO22+, DNAzyme-based sensors have the potential to greatly expand upon the range of metal ions able to be imaged using genetically-encoded proteins, allowing this class of sensors to be even more powerful in providing deeper understanding of the roles of metal ions in biology.
Supplementary Material
Acknowledgements
The work described in this article is supported by the U.S. National Institutes of Health (GM124316). We thank Dr. Sandra McMasters of the University of Illinois School of Chemical Sciences Cell Media Facility for assistance with cell culturing, the Institute for Genomic Biology Core Facility for assistance with confocal microscopy, and the Roy J. Carver Biotechnology for assistance with flow cytometry. We also thank Dr. Nitya Sai Reddy Satyavolu and Dr. Claire E. McGhee for their useful suggestions and support.
Footnotes
Supporting information for this article is given via a link at the end of the document
Contributor Information
Mengyi Xiong, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Collaborative Innovation Center for Chemistry and Molecular Medicine, Hunan University, Changsha 410082 (P.R. China); Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801 (USA).
Zhenglin Yang, Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, United States.
Ryan J. Lake, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801 (USA)
Junjie Li, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801 (USA).
Shanni Hong, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801 (USA).
Huanhuan Fan, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Collaborative Innovation Center for Chemistry and Molecular Medicine, Hunan University, Changsha 410082 (P.R. China); Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801 (USA).
Xiao-Bing Zhang, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Collaborative Innovation Center for Chemistry and Molecular Medicine, Hunan University, Changsha 410082 (P.R. China).
Yi Lu, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801 (USA); Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, United States.
References
- [1] a).Que EL, Domaille DW, Chang CJ, Chem. Rev 2008, 108, 1517–1549; [DOI] [PubMed] [Google Scholar]; b) Qian X, Xu Z, Chem. Soc. Rev 2015, 44, 4487–4493. [DOI] [PubMed] [Google Scholar]
- [2] a).Jiang P, Guo Z, Coord. Chem. Rev 2004, 248, 205–229; [Google Scholar]; b) Kikuchi K, Komatsu K, Nagano T, Curr. Opin. Chem. Biol 2004, 8, 182–191; [DOI] [PubMed] [Google Scholar]; c) Taki M, Wolford JL, O’Halloran TV, J. Am. Chem. Soc 2004, 126, 712–713; [DOI] [PubMed] [Google Scholar]; d) Komatsu K, Urano Y, Kojima H, Nagano T, J. Am. Chem. Soc 2007, 129, 13447–13454; [DOI] [PubMed] [Google Scholar]; e) Kennedy DP, Kormos CM, Burdette SC, J. Am. Chem. Soc 2009, 131, 8578–8586; [DOI] [PubMed] [Google Scholar]; f) McRae R, Bagchi P, Sumalekshmy S, Fahrni CJ, Chem. Rev 2009, 109, 4780–4827; [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Nolan EM, Lippard SJ, Acc. Chem. Res 2009, 42, 193–203; [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Mikata Y, Kawata K, Takeuchi S, Nakanishi K, Konno H, Itami S, Yasuda K, Tamotsu S, Burdette SC, Dalton Trans. 2014, 43, 10751–10759; [DOI] [PubMed] [Google Scholar]; i) Qiu L, Zhu C, Chen H, Hu M, He W, Guo Z, Chem. Commun 2014, 50, 4631–4634; [DOI] [PubMed] [Google Scholar]; j) Sareen D, Kaur P, Singh K, Coord. Chem. Rev 2014, 265, 125–154; [Google Scholar]; k) Cotruvo JA Jr., Aron AT, Ramos-Torres KM, Chang CJ, Chem. Soc. Rev 2015, 44, 4400–4414; [DOI] [PMC free article] [PubMed] [Google Scholar]; l) Aron AT, Loehr MO, Bogena J, Chang CJ, J. Am. Chem. Soc 2016, 138, 14338–14346; [DOI] [PMC free article] [PubMed] [Google Scholar]; m) Kaur N, Kaur P, Singh K, Sensor Actuat. B-Chem 2016, 229, 499–505; [Google Scholar]; n) Hirata T, Terai T, Yamamura H, Shirnonishi M, Komatsu T, Hanaoka K, Ueno T, Imaizumi Y, Nagano T, Urano Y, Anal. Chem 2016, 88, 2693–2700; [DOI] [PubMed] [Google Scholar]; o) Jia S, Ramos-Torres KM, Kolemen S, Ackerman CM, Chang CJ, ACS Chem. Biol 2018, 13, 1844–1852; [DOI] [PMC free article] [PubMed] [Google Scholar]; p) Bourassa D, Elitt CM, McCallum AM, Sumalekshmy S, McRae RL, Morgan MT, Siegel N, Perry JW, Rosenberg PA, Fahrni CJ, ACS Sensors 2018, 3, 458–467; [DOI] [PMC free article] [PubMed] [Google Scholar]; q) Goldberg JM, Wang F, Sessler CD, Vogler NW, Zhang DY, Loucks WH, Tzounopoulos T, Lippard SJ, J. Am. Chem. Soc 2018, 140, 2020–2023; [DOI] [PMC free article] [PubMed] [Google Scholar]; r) Matsui Y, Mizukami S, Kikuchi K, Chem. Lett 2018, 47, 23–26. [Google Scholar]
- [3].Huang X, Meng J, Dong Y, Cheng Y, Zhu C, Polymer 2010, 51, 3064–3067. [Google Scholar]
- [4] a).Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikurak M, Tsien RY, Nature 1997, 388, 882–887; [DOI] [PubMed] [Google Scholar]; b) Outten CE, O’Halloran TV, Science 2001, 292, 2488–2492; [DOI] [PubMed] [Google Scholar]; c) Chen P, He C, J. Am. Chem. Soc 2004, 126, 728–729; [DOI] [PubMed] [Google Scholar]; d) Palmer AE, Jin C, Reed JC, Tsien RY, Proc. Natl. Acad. Sci. USA 2004, 101, 17404–17409; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Van Dongen EMWM, Dekkers LM, Spijker K, Meijer EW, Klomp LWJ, Merkx M, J. Am. Chem. Soc 2006, 128, 10754–10762; [DOI] [PubMed] [Google Scholar]; f) Wegner SV, Okesli A, Chen P, He C, J. Am. Chem. Soc 2007, 129, 3474–3475; [DOI] [PubMed] [Google Scholar]; g) Wallace DJ, Borgloh S, Astori S, Yang Y, Bausen M, Kugler S, Palmer AE, Tsien RY, Sprengel R, Kerr JND, Denk W, Hasan MT, Nat. Methods 2008, 5, 797–804; [DOI] [PubMed] [Google Scholar]; h) Qin Y, Dittmer PJ, Park JG, Jansen KB, Palmer AE, Proc. Natl. Acad. Sci. USA 2011, 108, 7351–7356; [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Lindenburg LH, Vinkenborg JL, Oortwijn J, Aper SJA, Merkx M, Plos One 2013, 8, e82009; [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Carter KP, Young AM, Palmer AE, Chem. Rev 2014, 114, 4564–4601; [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Hessels AM, Merkx M, Metallomics 2015, 7, 258–266; [DOI] [PubMed] [Google Scholar]; l) Shen Y, Wu SY, Rancic V, Aggarwal A, Qian Y, Miyashita SI, Ballanyi K, Campbell RE, Dong M, Commun. Biol 2019, 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Park JG, Qin Y, Galati DF, Palmer AE, ACS Chem. Biol 2012, 7, 1636–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Koldenkova VP, Matsuda T, Nagai T, J. Biomed. Opt 2015, 20, 101203. [DOI] [PubMed] [Google Scholar]
- [7] a).Breaker RR, Joyce GF, Chem. Biol 1994, 1, 223–229; [DOI] [PubMed] [Google Scholar]; b) Feldman AR, Sen D, J. Mol. Biol 2001, 313, 283–294; [DOI] [PubMed] [Google Scholar]; c) Breaker RR, Emilsson GM, Lazarev D, Nakamura S, Puskarz IJ, Roth A, Sudarsan N, RNA 2003, 9, 949–957; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Breaker RR, Joyce GF, Chem. Biol 2014, 21, 1059–1065; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Liu J, Cao Z, Lu Y, Chem. Rev 2009, 109, 1948–1998; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Liu M, Chang D, Li Y, Acc. Chem. Res 2017, 50, 2273–2283. [DOI] [PubMed] [Google Scholar]
- [8] a).Santoro SW, Joyce GF, Proc. Natl. Acad. Sci. USA 1997, 94, 4262–4266; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Achenbach JC, Chiuman W, Cruz RPG, Li Y, Curr. Pharm. Biotechnol 2004, 5, 321–336; [DOI] [PubMed] [Google Scholar]; c) Torabi SF, Wu P, McGhee CE, Chen L, Hwang K, Zheng N, Cheng J, Lu Y, Proc. Natl. Acad. Sci. USA 2015, 112, 5903–5908; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zhang JJ, Cheng FF, Li JJ, Zhu J-J, Lu Y, Nano Today, 2016, 11, 309–329; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) McGhee CE, Loh KY, Lu Y, Curr. Opin. Biotechnol 2017, 45, 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9] a).Li J, Lu Y, J. Am. Chem. Soc 2000, 122, 10466–10467; [Google Scholar]; b) Wu P, Hwang K, Lan T, Lu Y, J. Am. Chem. Soc 2013, 135, 5254–5257; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hwang K, Wu P, Kim T, Lei L, Tian S, Wang Y, Lu Y, Angew. Chem. Int. Ed 2014, 53, 13798–13802; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Saran R, Liu J, Anal. Chem 2016, 88, 4014–4020; [DOI] [PubMed] [Google Scholar]; e) Zhou W, Zhang Y, Ding J, Liu J, ACS Sensors 2016, 1, 600–606; [Google Scholar]; f) Zhou W, Saran R, Liu J, Chem. Rev 2017, 117, 8272–8325; [DOI] [PubMed] [Google Scholar]; g) Xiang Y, Wang Z, Xing H, Wong NY, Lu Y, Anal. Chem 2010, 82, 4122–4129; [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Yang Z, Loh KY, Chu YT, Feng R, Satyavolu NSR, Xiong M, Huynh SMN, Hwang K, Li L, Xing H, Zhang X, Chelma YR, Gruebele M, Lu Y, J. Am. Chem. Soc 2018, 140, 17656–17665; [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Lake R, Yang Z, Zhang J, Lu Y, Acc. Chem. Res 2019, DOI: 10.1021/acs.accounts.9b00419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, McKeown MR, Wiedenmann J, Davidson MW, Schnitzer MJ, Tsien RY, Lin MZ, Nat. Methods 2012, 9, 1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11] a).Santiago FS, Lowe HC, Kavurma MM, Chesterman CN, Baker A, Atkins DG, Khachigian LM, Nat. Med 1999, 5, 1264–1269; [DOI] [PubMed] [Google Scholar]; b) Garn H, Renz H, Eur. J. Immunol 2017, 47, 22–30; [DOI] [PubMed] [Google Scholar]; c) Yehl K, Joshi JP, Greene BL, Dyer RB, Nahta R, Salaita K, ACS Nano 2012, 6, 9150–9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Young DD, Lively MO, Deiters A, J. Am. Chem. Soc 2010, 132, 6183–6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13] a).Fan H, Zhao Z, Yan G, Zhang X, Yang C, Meng H, Chen Z, Liu H, Tan W, Angew. Chem. Int. Ed 2015, 54, 4801–4805; [DOI] [PubMed] [Google Scholar]; b) Kim JE, Yoon S, Choi BR, Kim KP, Cho YH, Jung W, Kim DW, Oh S, Kim DE, Leukemia 2013, 27, 1650–1658; [DOI] [PubMed] [Google Scholar]; c) Wang DY, Lai BHY, Sen D, J. Mol. Biol 2002, 318, 33–43. [DOI] [PubMed] [Google Scholar]
- [14].Cairns MJ, Nucleic Acids Res. 2003, 31, 2883–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cairns MJ, Hopkins TM, Witherington C, Sun LQ, Antisense Nucleic A 2000, 10, 323–332. [DOI] [PubMed] [Google Scholar]
- [16].Fokina AA, Meschaninova MI, Durfort T, Venyaminova AG, Francois JC, Biochemistry 2012, 51, 2181–2191. [DOI] [PubMed] [Google Scholar]
- [17].Sou SN, Polizzi KM, Kontoravdi C, Adv. Biosci. Biotech 2013, 04, 1013–1019. [Google Scholar]
- [18].Hwang K, Mou Q, Lake R, Xiong M, Holland B, Lu Y, Inorg. Chem 2019, 58, 13696–13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19] a).Li J, Zheng W, Kwon A, Lu Y, Nucleic Acids Res. 2000, 28, 481–488; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lu Y, Liu J, Li J, Bruesehoff PJ, Pavot CM-B, Brown AK, Biosen. Bioeletron 18, 529–540 (2003); [DOI] [PubMed] [Google Scholar]; c) Bonaccio M, Credali A, Peracchi A, Nucleic Acids Res. 2004, 32, 916–925; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Schlosser K, Gu J, Sule L, Li Y, Nucleic Acids Res. 2008, 36, 1472–1481; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Wang F, Saran R, Liu J, Bioorg. Med. Chem. Lett 2015, 25, 1460–1463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.