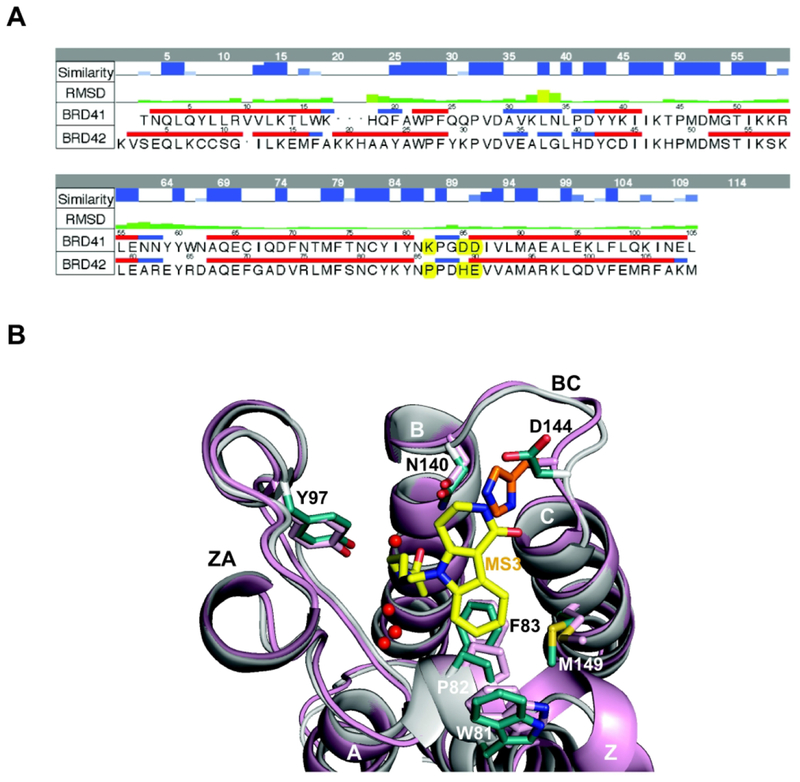
Figure 7. Selectivity binding of Olinone to BRD4 BrD1 versus BRD4 BrD2.
(A) Sequence alignment between the two BRD4 BrDs, BrD1 and BrD2 (Some amino acids that are different in both BrDs, including the Asp144 (BrD1) and His437 (BrD2) are highlighted in yellow). To align the residues numbering of both X-ray crystal structures, 59 units should be added to the figure residue numbering for BrD1 and 348 units for BrD2. The top two rows in the table represent the similarity and RMSD between the two structures, respectively. The alignment was made using MOE18. (B) Structural comparison of BRD4 BrD1 (gray, PDB ID 3UVW19) and BRD4 BrD2 (pink, PDB ID 2OUO19,50) X-ray crystal structures, showing steric clash between MS3 (yellow) and His437 in BRD4 BrD2 (orange) rendered using PyMOL program.34 Side chains of key amino acid residues at the ligand-binding site in the protein are color-coded by atom type. Water molecules in the ligand-binding site are shown as red spheres.