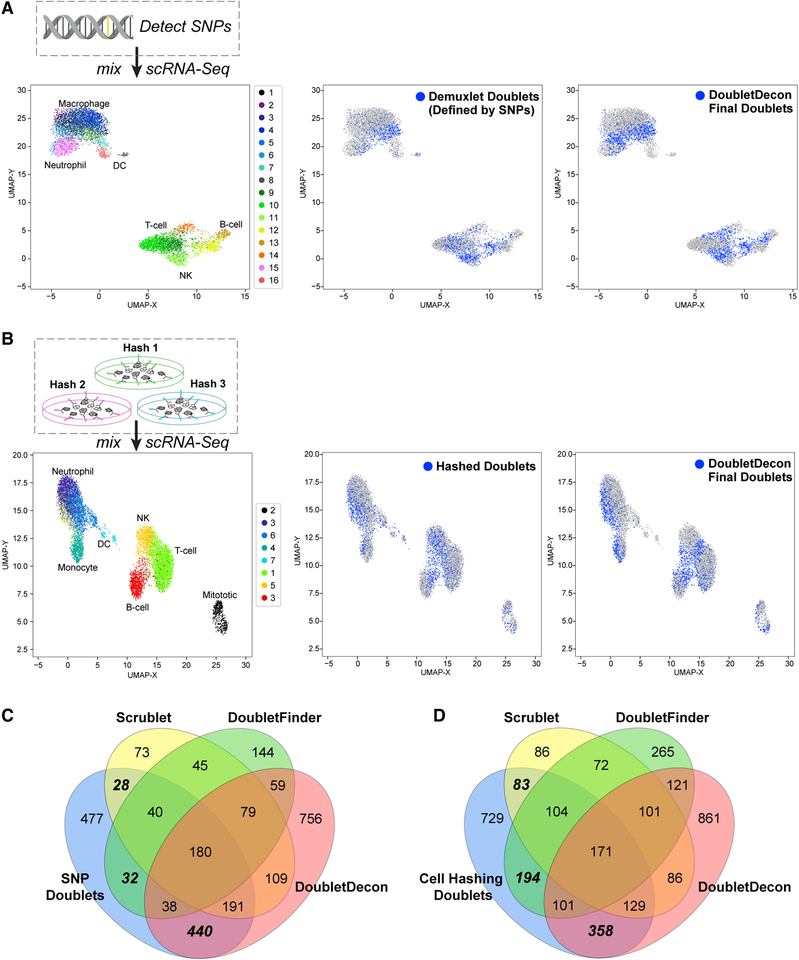
Figure 4. Detection of Experimentally Validated Doublets from Peripheral Blood Mononuclear Cells (PBMCs).
(A and B) The analysis schema is shown for the evaluation of DoubletDecon on in silico identified doublet cell profiles obtained from the (A) Dexmulet software and (B) the Cell Hashing protocol. Demuxlet identifies cells with a combination of genomic variants associated with the eight profiled single-cell donors to find cellular bar codes with hybrid genotype profiles, whereas Cell Hashing selectively labels all cells from a single sample (donor) using different oligonucleotides conjugated to a common antibody. (Left) A Uniform Manifold Approximation and Projection (UMAP) plot of the de novo clusters obtained from analysis with ICGS. (Middle) UMAP projection of Demuxlet called doublets are indicated in blue. (Right) UMAP projections of DoubletDecon-classified doublets are highlighted in blue. Labels for each cell population were independently derived through ICGS version 2.0 using a published database of hematopeotic and immune markers via GO-Elite gene set enrichment analysis (Hay et al., 2018).
(C and D) Venn diagrams representing the number of overlapping doublet predictions from the software packages DoubletDecon, Scrublet, and DoubletFinder on two previously published datasets of overloaded donor PBMCs using the (C) Demuxlet or (D) Cell Hashing protocols using the same filtered datasets described above. Hashing doublets, doublets defined from distinct hashtag oligo (HTO). If two or more HTOs had >20% of the total hashtag reads, they were considered multiplets (4,200 out of the initial total 12,000 cellular bar codes). Demuxlet doublets, doublets identified by Kang et al. (2018) using the software Demuxlet.