Figure 5. Empirical Removal of Confounding Doublet-Cell Populations for Unsupervised Subtype Detection.
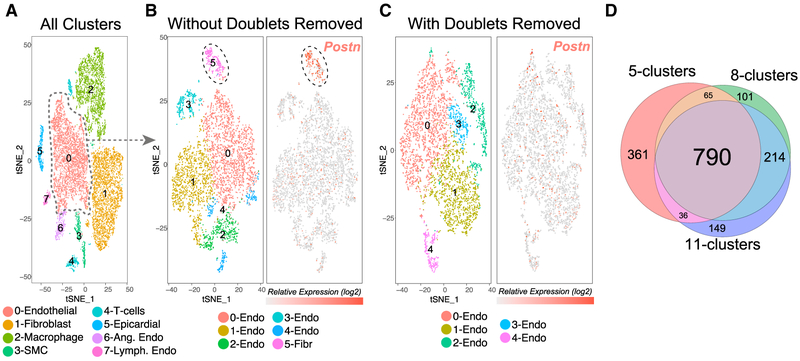
(A) t-SNE visualization of the predominant cell populations identified from Seurat of ~13,000 heart cells collected via Drop-Seq. (Left panel) Cell-type predictions are based on established heart marker genes (literature) and gene set enrichment in the software GO-Elite (cellular biomarker database). (Right panel) DoubletDecon doublet predictions overlaid on top of the Seurat t-SNE plot, localized to the periphery of the major Seurat clusters. The dashed circle highlights endothelial-specific predicted doublets adjacent to fibroblasts.
(B and C) Secondary analysis of all Seurat-identified endothelial cells with (B) all doublets included and (C) doublets excluded with DoubletDecon prior to clustering. The left panel indicates distinct endothelial cell clusters with the doublet-enriched fibroblast cells highlighted (dashed circle), while the right panel visualizes expression of a fibroblast-specific marker.
(D) Venn diagram of DoubletDecon doublet predictions with three sepearte Seurat clustering resolutions of the entire heart dataset. The numbers of doublets identified were 1,251 (5 clusters), 1,170 (8 clusters), and 1,189 (11 clusters), with 790 (63%) in common.